UniProt ID: P0C8G7 (None-None) Perivitellin-2 31 kDa subunit
UniProt ID: P0C8G6 (None-None) Perivitellin-2 67 kDa subunit
|
|
|

|
| Sample: |
Perivitellin-2 31 kDa subunit tetramer, 126 kDa Pomacea canaliculata protein
Perivitellin-2 67 kDa subunit tetramer, 250 kDa Pomacea canaliculata protein
|
| Buffer: |
20 mM Tris-HCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2014 Jun 10
|
Apple Snail Perivitellin Precursor Properties Help Explain Predators' Feeding Behavior.
Physiol Biochem Zool 90(4):461-470 (2017)
Cadierno MP, Dreon MS, Heras H
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
294 |
nm3 |
|
|
UniProt ID: Q6NUC7 (2-145) Nucleoplasmin core + A2
UniProt ID: Q6AZJ8 (2-130) Histone H2A (ΔAla127)
UniProt ID: P02281 (5-126) Histone H2B 1.1 (Ser33Thr)
|
|
|

|
| Sample: |
Nucleoplasmin core + A2 pentamer, 81 kDa Xenopus laevis protein
Histone H2A (ΔAla127) pentamer, 69 kDa Xenopus laevis protein
Histone H2B 1.1 (Ser33Thr) pentamer, 67 kDa Xenopus laevis protein
|
| Buffer: |
20 mM Tris. 150 mM NaCl, 1 mM EDTA, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2016 Jan 7
|
Dynamic intramolecular regulation of the histone chaperone nucleoplasmin controls histone binding and release.
Nat Commun 8(1):2215 (2017)
Warren C, Matsui T, Karp JM, Onikubo T, Cahill S, Brenowitz M, Cowburn D, Girvin M, Shechter D
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
402 |
nm3 |
|
|
UniProt ID: P07101 (1-497) Tyrosine hydroxylase, isoform 1
|
|
|
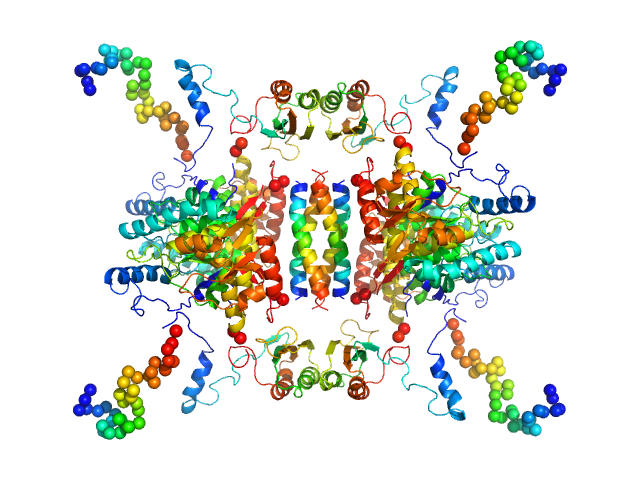
|
| Sample: |
Tyrosine hydroxylase, isoform 1 tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES 200 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 14
|
Stable preparations of tyrosine hydroxylase provide the solution structure of the full-length enzyme.
Sci Rep 6:30390 (2016)
Bezem MT, Baumann A, Skjærven L, Meyer R, Kursula P, Martinez A, Flydal MI
|
| RgGuinier |
4.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
520 |
nm3 |
|
|
UniProt ID: Q14896 (641-964) Cardiac myosin binding protein-C: domains C5-C6-C7
|
|
|
|
| Sample: |
Cardiac myosin binding protein-C: domains C5-C6-C7 monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 250 mM NaCl, 2 mM TCEP, 0.02% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 18
|
Clinically Linked Mutations in the Central Domains of Cardiac Myosin-Binding Protein C with Distinct Phenotypes Show Differential Structural Effects.
Structure 24(1):105-115 (2016)
Nadvi NA, Michie KA, Kwan AH, Guss JM, Trewhella J
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
UniProt ID: Q6GEY1 (None-None) Thiaminase type II enzyme
|
|
|
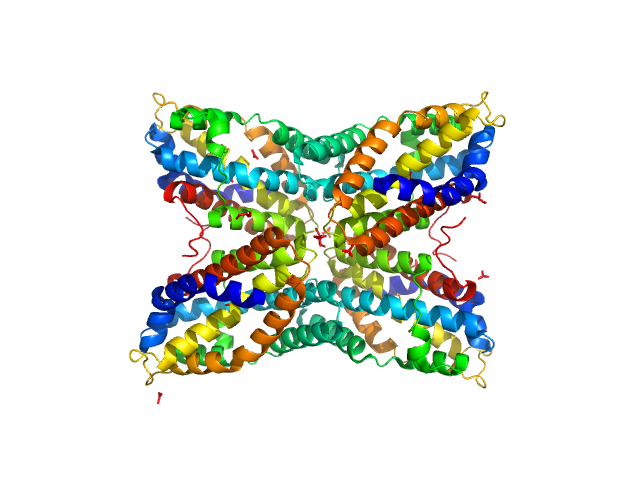
|
| Sample: |
Thiaminase type II enzyme tetramer, 107 kDa Staphylococcus aureus protein
|
| Buffer: |
100 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 11
|
Staphylococcus aureus thiaminase II: oligomerization warrants proteolytic protection against serine proteases.
Acta Crystallogr D Biol Crystallogr 69(Pt 12):2320-9 (2013)
Begum A, Drebes J, Kikhney A, Müller IB, Perbandt M, Svergun D, Wrenger C, Betzel C
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
UniProt ID: P0AEU7 (21-161) Periplasmic holdase chaperone protein Skp
|
|
|
|
| Sample: |
Periplasmic holdase chaperone protein Skp trimer, 47 kDa Escherichia coli protein
|
| Buffer: |
25 mM HEPES 150 mM NaCl 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 24
|
A Spring-Loaded Mechanism Governs the Clamp-like Dynamics of the Skp Chaperone.
Structure 25(7):1079-1088.e3 (2017)
Holdbrook DA, Burmann BM, Huber RG, Petoukhov MV, Svergun DI, Hiller S, Bond PJ
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
UniProt ID: I3NIC8 (1-141) SycH putative yopH targeting protein
UniProt ID: P08538 (1-129) Tyrosine-protein phosphatase YopH
|
|
|
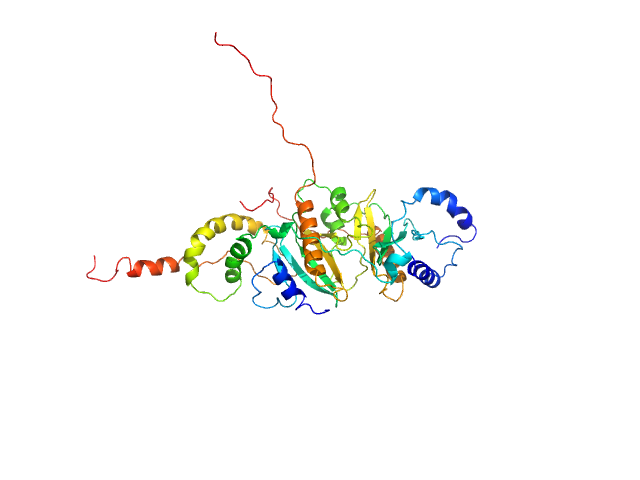
|
| Sample: |
SycH putative yopH targeting protein dimer, 32 kDa Yersinia pseudotuberculosis protein
Tyrosine-protein phosphatase YopH monomer, 14 kDa Yersinia pseudotuberculosis protein
|
| Buffer: |
50 mM HEPES 2mM TCEP, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Aug 1
|
Global Disordering in Stereo-Specific Protein Association
Biophysical Journal 112(3):33a (2017)
Gupta A, Reinartz I, Spilotros A, Jonna V, Hofer A, Svergun D, Schug A, Wolf-Watz M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
UniProt ID: Q46703 (None-None) Escherichia coli TraE protein (VirB8 homolog)
|
|
|

|
| Sample: |
Escherichia coli TraE protein (VirB8 homolog) hexamer, 171 kDa Escherichia coli protein
|
| Buffer: |
50 mM sodium phosphate 300 mM NaCl 40 mM imidazole 0.15 % octyl glucose neopentyl glycol (OGNG), pH: 7.4 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Jun 2
|
VirB8 homolog TraE from plasmid pKM101 forms a hexameric ring structure and interacts with the VirB6 homolog TraD.
Proc Natl Acad Sci U S A 115(23):5950-5955 (2018)
Casu B, Mary C, Sverzhinsky A, Fouillen A, Nanci A, Baron C
|
| RgGuinier |
4.4 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
360 |
nm3 |
|
|
UniProt ID: Q9JYV5 (415-591) Iron-regulated protein FrpC (amino acids 415-591)
|
|
|
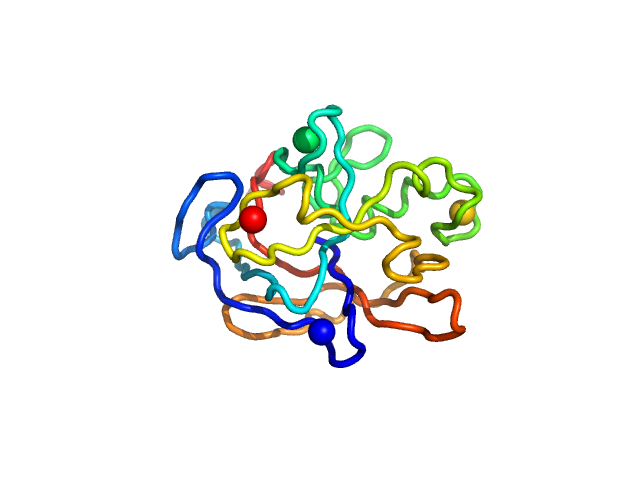
|
| Sample: |
Iron-regulated protein FrpC (amino acids 415-591) monomer, 19 kDa Neisseria meningitidis protein
|
| Buffer: |
5 mM Tris 50 mM NaCl 10 mM CaCl2 0.1% NaN3, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2013 Oct 3
|
Self-processing module of Iron-regulated protein FrpC
Vojtěch Kubáň
|
| RgGuinier |
1.7 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
UniProt ID: Q7BCK4 (None-None) Outer membrane protein IcsA (53-758)
|
|
|

|
| Sample: |
Outer membrane protein IcsA (53-758) monomer, 72 kDa Shigella flexneri protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 10 mM CaCl2 3% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 May 18
|
The Shigella Virulence Factor IcsA Relieves N-WASP Autoinhibition by Displacing the Verprolin Homology/Cofilin/Acidic (VCA) Domain.
J Biol Chem 292(1):134-145 (2017)
Mauricio RP, Jeffries CM, Svergun DI, Deane JE
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
103 |
nm3 |
|
|