UniProt ID: Q8DNZ9 (1-447) Lipid II isoglutaminyl synthase (glutamine-hydrolyzing) subunit MurT
UniProt ID: Q8DNZ8 (None-None) Lipid II isoglutaminyl synthase (glutamine-hydrolyzing) subunit GatD
|
|
|
|
| Sample: |
Lipid II isoglutaminyl synthase (glutamine-hydrolyzing) subunit MurT monomer, 52 kDa Streptococcus pneumoniae (strain … protein
Lipid II isoglutaminyl synthase (glutamine-hydrolyzing) subunit GatD monomer, 29 kDa Streptococcus pneumoniae (strain … protein
|
| Buffer: |
50 mM Hepes, 10 mM MgCl2, 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 23
|
Unravelling the reaction mechanism of glutamate amidation in Staphylococcus aureus peptidoglycan
PhD thesis, NOVA University Lisbon - (2022)
Francisco Miguel Piçarra Leisico, Mertens HD
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
UniProt ID: A0A0H2WZQ7 (2-437) Lipid II isoglutaminyl synthase (glutamine-hydrolyzing) subunit MurT
UniProt ID: A0A0H2WZ38 (1-243) Lipid II isoglutaminyl synthase (glutamine-hydrolyzing) subunit GatD
|
|
|
|
| Sample: |
Lipid II isoglutaminyl synthase (glutamine-hydrolyzing) subunit MurT monomer, 49 kDa Staphylococcus aureus (strain … protein
Lipid II isoglutaminyl synthase (glutamine-hydrolyzing) subunit GatD monomer, 28 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
100 mM Tris-HCl, 500 mM NaCl, 10 mM MgCl2, pH: 8.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Nov 29
|
Unravelling the reaction mechanism of glutamate amidation in Staphylococcus aureus peptidoglycan
PhD thesis, NOVA University Lisbon - (2022)
Francisco Miguel Piçarra Leisico, Mertens HD
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
121 |
nm3 |
|
|
UniProt ID: Q9WZ31 (1-351) Cobalt/magnesium transport protein CorA
|
|
|
|
| Sample: |
Cobalt/magnesium transport protein CorA pentamer, 208 kDa Thermotoga maritima (strain … protein
|
| Buffer: |
20 mM Tris-DCl, 150 mM NaCl, 1 mM EDTA-NaOD, in 100% D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 Jun 23
|
Mg2+-dependent conformational equilibria in CorA and an integrated view on transport regulation.
Elife 11 (2022)
Johansen NT, Bonaccorsi M, Bengtsen T, Larsen AH, Tidemand FG, Pedersen MC, Huda P, Berndtsson J, Darwish T, Yepuri NR, Martel A, Pomorski TG, Bertarello A, Sansom M, Rapp M, Crehuet R, Schubeis T, Lindorff-Larsen K, Pintacuda G, Arleth L
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.9 |
nm |
|
|
UniProt ID: Q9WZ31 (1-351) Cobalt/magnesium transport protein CorA
|
|
|
|
| Sample: |
Cobalt/magnesium transport protein CorA pentamer, 208 kDa Thermotoga maritima (strain … protein
|
| Buffer: |
20 mM Tris-DCl, 150 mM NaCl, 40 mM MgCl2, in 100% D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 Jun 23
|
Mg2+-dependent conformational equilibria in CorA and an integrated view on transport regulation.
Elife 11 (2022)
Johansen NT, Bonaccorsi M, Bengtsen T, Larsen AH, Tidemand FG, Pedersen MC, Huda P, Berndtsson J, Darwish T, Yepuri NR, Martel A, Pomorski TG, Bertarello A, Sansom M, Rapp M, Crehuet R, Schubeis T, Lindorff-Larsen K, Pintacuda G, Arleth L
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.9 |
nm |
|
|
UniProt ID: Q9WZ31 (1-351) Cobalt/magnesium transport protein CorA
|
|
|
|
| Sample: |
Cobalt/magnesium transport protein CorA pentamer, 208 kDa Thermotoga maritima (strain … protein
|
| Buffer: |
20 mM Tris-DCl, 150 mM NaCl, 1 mM EDTA-NaOD, in 100% D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 Jun 23
|
Mg2+-dependent conformational equilibria in CorA and an integrated view on transport regulation.
Elife 11 (2022)
Johansen NT, Bonaccorsi M, Bengtsen T, Larsen AH, Tidemand FG, Pedersen MC, Huda P, Berndtsson J, Darwish T, Yepuri NR, Martel A, Pomorski TG, Bertarello A, Sansom M, Rapp M, Crehuet R, Schubeis T, Lindorff-Larsen K, Pintacuda G, Arleth L
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.2 |
nm |
|
|
UniProt ID: Q9WZ31 (1-351) Cobalt/magnesium transport protein CorA
|
|
|
|
| Sample: |
Cobalt/magnesium transport protein CorA pentamer, 208 kDa Thermotoga maritima (strain … protein
|
| Buffer: |
20 mM Tris-DCl, 150 mM NaCl, 40 mM MgCl2, in 100% D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 Jun 23
|
Mg2+-dependent conformational equilibria in CorA and an integrated view on transport regulation.
Elife 11 (2022)
Johansen NT, Bonaccorsi M, Bengtsen T, Larsen AH, Tidemand FG, Pedersen MC, Huda P, Berndtsson J, Darwish T, Yepuri NR, Martel A, Pomorski TG, Bertarello A, Sansom M, Rapp M, Crehuet R, Schubeis T, Lindorff-Larsen K, Pintacuda G, Arleth L
|
| RgGuinier |
4.9 |
nm |
| Dmax |
14.3 |
nm |
|
|
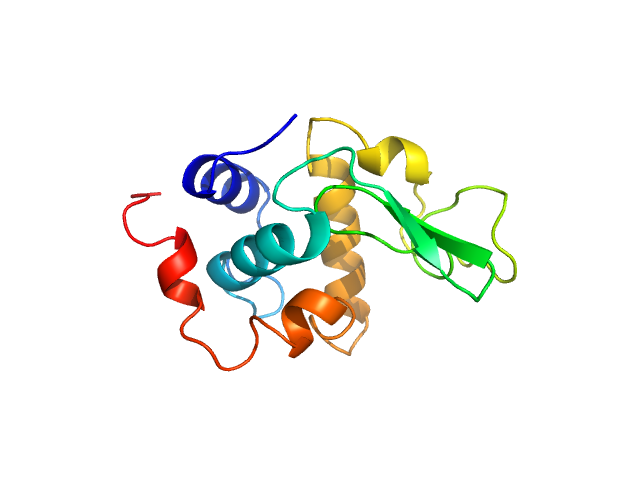
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.3 |
nm |
| Dmax |
4.2 |
nm |
|
|
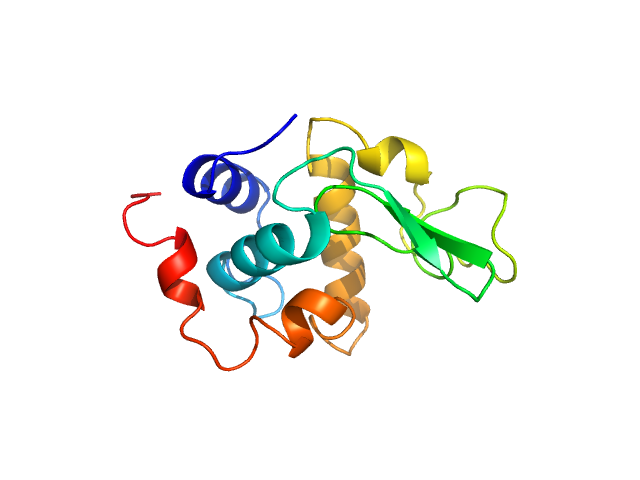
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
|
|
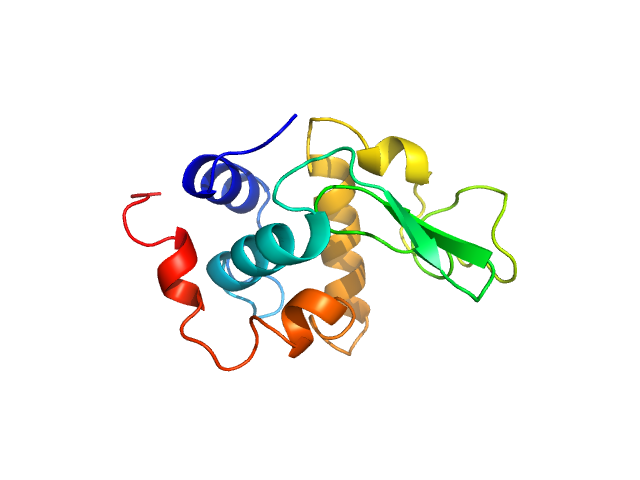
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.5 |
nm |
|
|
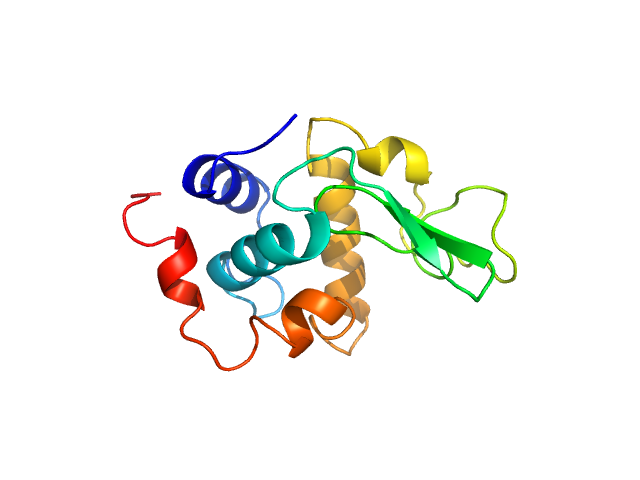
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
40 mM NaOAc pH 3.8, 150 mM NaCl, pH: 3.8 |
| Experiment: |
SAXS
data collected at X9A, National Synchrotron Light Source (NSLS) on 2014 May 2
|
Visualizing how inclusion of higher reciprocal space in SWAXS data analysis improves shape restoration of biomolecules: case of lysozyme.
J Biomol Struct Dyn :1-15 (2021)
Ashish
|
| RgGuinier |
1.4 |
nm |
| Dmax |
5.0 |
nm |
|
|