UniProt ID: P62508 (122-458) Estrogen-related receptor gamma
UniProt ID: None (None-None) Inverse repeat IR3 DNA
|
|
|

|
| Sample: |
Estrogen-related receptor gamma hexamer, 230 kDa Homo sapiens protein
Inverse repeat IR3 DNA trimer, 46 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1% (v/v) glycerol, and 1 mM CHAPS, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 3
|
Reconstruction of Quaternary Structure from X-ray Scattering by Equilibrium Mixtures of Biological Macromolecules
Biochemistry 52(39):6844-6855 (2013)
Petoukhov M, Billas I, Takacs M, Graewert M, Moras D, Svergun D
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.2 |
nm |
|
|
UniProt ID: P62508 (122-458) Estrogen-related receptor gamma
UniProt ID: None (None-None) Inverse repeat IR3 DNA
|
|
|

|
| Sample: |
Estrogen-related receptor gamma hexamer, 230 kDa Homo sapiens protein
Inverse repeat IR3 DNA trimer, 46 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1% (v/v) glycerol, and 1 mM CHAPS, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Mar 3
|
Reconstruction of Quaternary Structure from X-ray Scattering by Equilibrium Mixtures of Biological Macromolecules
Biochemistry 52(39):6844-6855 (2013)
Petoukhov M, Billas I, Takacs M, Graewert M, Moras D, Svergun D
|
| RgGuinier |
4.1 |
nm |
| Dmax |
12.8 |
nm |
|
|
UniProt ID: P0CY32 (2-166) GTP-binding domain of Ras-like protein 1
|
|
|
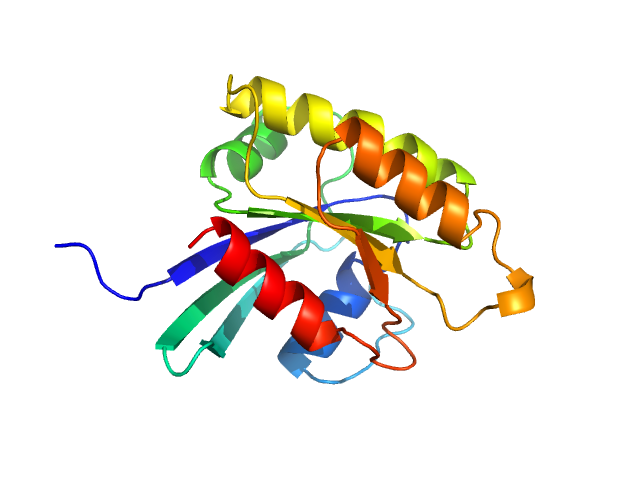
|
| Sample: |
GTP-binding domain of Ras-like protein 1 monomer, 19 kDa protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% glycerol, 3 mM DTT, 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 21
|
Pathogen-specific structural features of Candida albicans Ras1 activation complex: uncovering new antifungal drug targets.
mBio :e0063823 (2023)
Manso JA, Carabias A, Sárkány Z, de Pereda JM, Pereira PJB, Macedo-Ribeiro S
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.1 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
UniProt ID: P0CY32 (2-290) Ras-like protein 1
|
|
|
|
| Sample: |
Ras-like protein 1 monomer, 33 kDa Candida albicans protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 5% glycerol, 3 mM DTT, 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 21
|
Pathogen-specific structural features of Candida albicans Ras1 activation complex: uncovering new antifungal drug targets.
mBio :e0063823 (2023)
Manso JA, Carabias A, Sárkány Z, de Pereda JM, Pereira PJB, Macedo-Ribeiro S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.1 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
UniProt ID: P43069 (860-1333) Cell division control protein 25
|
|
|
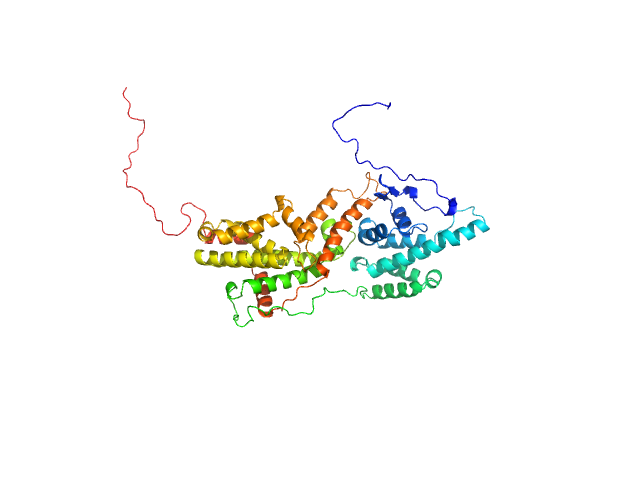
|
| Sample: |
Cell division control protein 25 monomer, 55 kDa Candida albicans (strain … protein
|
| Buffer: |
20 mM Na-phosphate, 150 mM NaCl, 5% glycerol, 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 21
|
Pathogen-specific structural features of Candida albicans Ras1 activation complex: uncovering new antifungal drug targets.
mBio :e0063823 (2023)
Manso JA, Carabias A, Sárkány Z, de Pereda JM, Pereira PJB, Macedo-Ribeiro S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
UniProt ID: P0CY32 (2-166) GTP-binding domain of Ras-like protein 1
UniProt ID: P43069 (860-1333) Cell division control protein 25
|
|
|
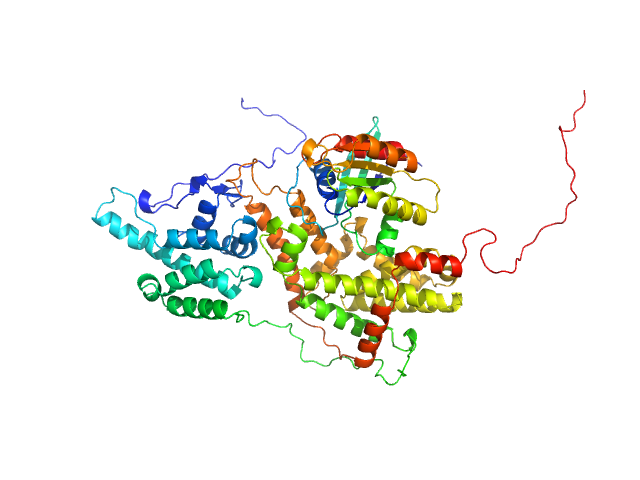
|
| Sample: |
GTP-binding domain of Ras-like protein 1 monomer, 19 kDa protein
Cell division control protein 25 monomer, 55 kDa Candida albicans (strain … protein
|
| Buffer: |
20 mM Na-phosphate, 150 mM NaCl, 1 mM EDTA, 5% glycerol, 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 21
|
Pathogen-specific structural features of Candida albicans Ras1 activation complex: uncovering new antifungal drug targets.
mBio :e0063823 (2023)
Manso JA, Carabias A, Sárkány Z, de Pereda JM, Pereira PJB, Macedo-Ribeiro S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
UniProt ID: P0CY32 (2-290) Ras-like protein 1
UniProt ID: P43069 (860-1333) Cell division control protein 25
|
|
|
|
| Sample: |
Ras-like protein 1 monomer, 33 kDa Candida albicans protein
Cell division control protein 25 monomer, 55 kDa Candida albicans (strain … protein
|
| Buffer: |
20 mM Na-phosphate, 150 mM NaCl, 1 mM EDTA, 5% glycerol, 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 21
|
Pathogen-specific structural features of Candida albicans Ras1 activation complex: uncovering new antifungal drug targets.
mBio :e0063823 (2023)
Manso JA, Carabias A, Sárkány Z, de Pereda JM, Pereira PJB, Macedo-Ribeiro S
|
| RgGuinier |
4.0 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
149 |
nm3 |
|
|
UniProt ID: O43186 (39-98) Cone-rod homeobox protein
|
|
|
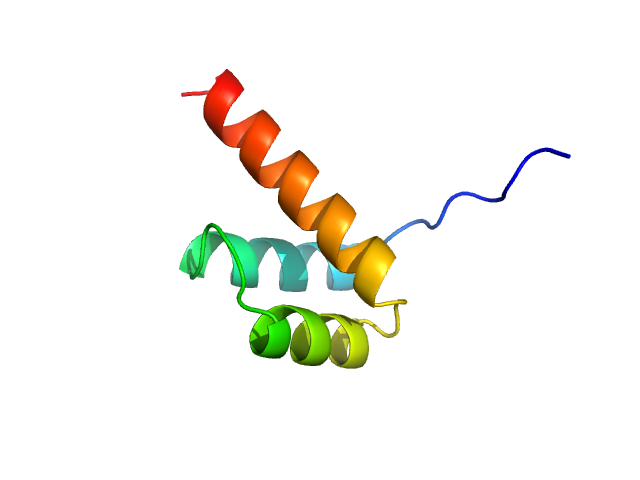
|
| Sample: |
Cone-rod homeobox protein monomer, 7 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate,100 mM NaCl, and 5 mM imidazole, pH: 7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Jun 15
|
Structural and functional analysis of the human Cone-rod homeobox transcription factor.
Proteins (2022)
Clanor PB, Buchholz C, Hayes JE, Friedman MA, White AM, Enke RA, Berndsen CE
|
| RgGuinier |
1.2 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
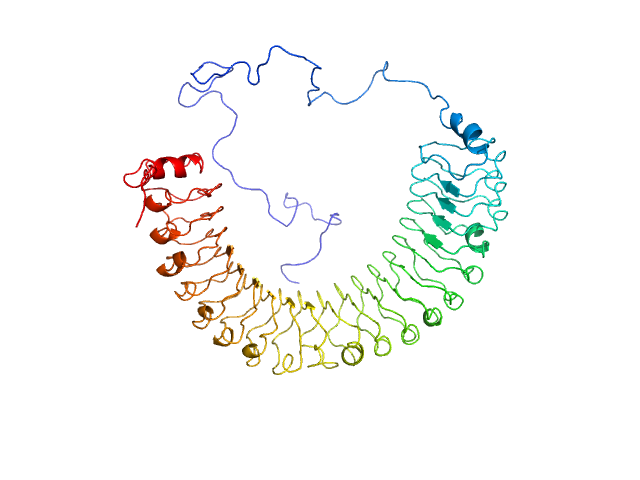
UniProt ID: Q9UQ13 (2-582) Leucine-rich repeat protein SHOC-2
|
|
|
|
| Sample: |
Leucine-rich repeat protein SHOC-2 monomer, 65 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris pH 7.5, 100 mM NaCl, 1 mM TCEP, 1 mM MnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jun 20
|
Structural basis for SHOC2 modulation of RAS signalling.
Nature (2022)
Liau NPD, Johnson MC, Izadi S, Gerosa L, Hammel M, Bruning JM, Wendorff TJ, Phung W, Hymowitz SG, Sudhamsu J
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
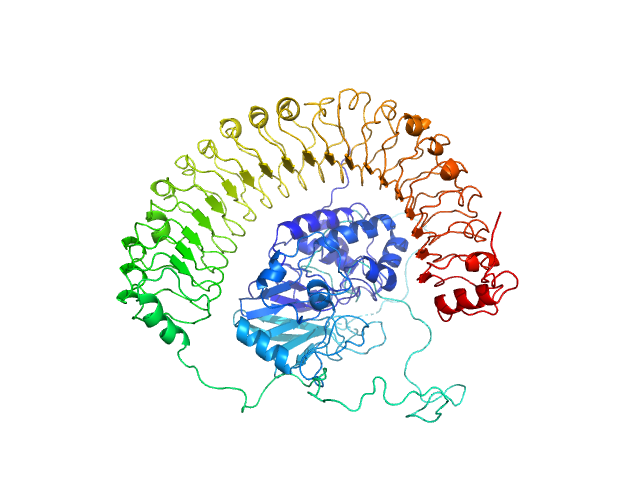
UniProt ID: Q9UQ13 (2-582) Leucine-rich repeat protein SHOC-2
UniProt ID: P36873 (1-323) Serine/threonine-protein phosphatase PP1-gamma catalytic subunit
|
|
|
|
| Sample: |
Leucine-rich repeat protein SHOC-2 monomer, 65 kDa Homo sapiens protein
Serine/threonine-protein phosphatase PP1-gamma catalytic subunit monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris pH 7.5, 100 mM NaCl, 1 mM TCEP, 1 mM MnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 20
|
Structural basis for SHOC2 modulation of RAS signalling.
Nature (2022)
Liau NPD, Johnson MC, Izadi S, Gerosa L, Hammel M, Bruning JM, Wendorff TJ, Phung W, Hymowitz SG, Sudhamsu J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
132 |
nm3 |
|
|