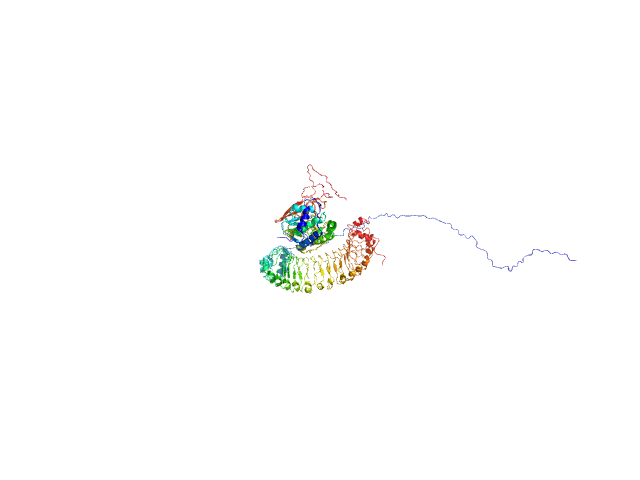
UniProt ID: Q9UQ13 (2-582) Leucine-rich repeat protein SHOC-2
UniProt ID: P36873 (1-323) Serine/threonine-protein phosphatase PP1-gamma catalytic subunit
UniProt ID: P01116 (2-150) GTPase KRas
|
|
|
|
| Sample: |
Leucine-rich repeat protein SHOC-2 monomer, 65 kDa Homo sapiens protein
Serine/threonine-protein phosphatase PP1-gamma catalytic subunit monomer, 37 kDa Homo sapiens protein
GTPase KRas monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris pH 7.5, 100 mM NaCl, 1 mM TCEP, 1 mM MnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 20
|
Structural basis for SHOC2 modulation of RAS signalling.
Nature (2022)
Liau NPD, Johnson MC, Izadi S, Gerosa L, Hammel M, Bruning JM, Wendorff TJ, Phung W, Hymowitz SG, Sudhamsu J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
156 |
nm3 |
|
|
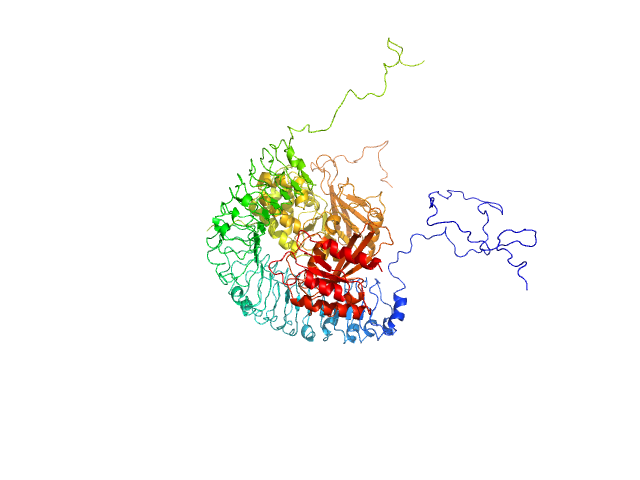
UniProt ID: Q9UQ13 (2-582) Leucine-rich repeat protein SHOC-2
UniProt ID: P36873 (1-323) Serine/threonine-protein phosphatase PP1-gamma catalytic subunit
UniProt ID: O14807 (1-208) Ras-related protein M-Ras
|
|
|
|
| Sample: |
Leucine-rich repeat protein SHOC-2 monomer, 65 kDa Homo sapiens protein
Serine/threonine-protein phosphatase PP1-gamma catalytic subunit monomer, 37 kDa Homo sapiens protein
Ras-related protein M-Ras monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris pH 7.5, 100 mM NaCl, 1 mM TCEP, 1 mM MnCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 20
|
Structural basis for SHOC2 modulation of RAS signalling.
Nature (2022)
Liau NPD, Johnson MC, Izadi S, Gerosa L, Hammel M, Bruning JM, Wendorff TJ, Phung W, Hymowitz SG, Sudhamsu J
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.4 |
nm |
| VolumePorod |
215 |
nm3 |
|
|
UniProt ID: M1QZ58 (1-987) TnpA transposase
|
|
|

|
| Sample: |
TnpA transposase dimer, 234 kDa Bacillus thuringiensis serovar … protein
|
| Buffer: |
50 mM HEPES, 200 mM NaCl, 100 mM L-Arg HCL, pH: 7.9 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Nov 2
|
AFM-based force spectroscopy unravels stepwise formation of the DNA transposition complex in the widespread Tn3 family mobile genetic elements.
Nucleic Acids Res (2023)
Fernandez M, Shkumatov AV, Liu Y, Stulemeijer C, Derclaye S, Efremov RG, Hallet B, Alsteens D
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
480 |
nm3 |
|
|
UniProt ID: A0A0D1DWZ5 (4-792) RNA recognition motif (RRM)-containing protein 4
|
|
|
|
| Sample: |
RNA recognition motif (RRM)-containing protein 4 monomer, 111 kDa Ustilago maydis protein
|
| Buffer: |
20 mM Hepes, 200 mM NaCl, 1 mM βME, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 30
|
A MademoiseLLE domain binding platform links the key RNA transporter to endosomes
PLOS Genetics 18(6):e1010269 (2022)
Devan S, Schott-Verdugo S, Müntjes K, Bismar L, Reiners J, Hachani E, Schmitt L, Höppner A, Smits S, Gohlke H, Feldbrügge M, Mitchell A
|
| RgGuinier |
8.8 |
nm |
| Dmax |
30.7 |
nm |
| VolumePorod |
587 |
nm3 |
|
|
UniProt ID: A0A0D1DWZ5 (421-792) RNA recognition motif (RRM)-containing protein 4 NT4
|
|
|
|
| Sample: |
RNA recognition motif (RRM)-containing protein 4 NT4 monomer, 40 kDa Ustilago maydis protein
|
| Buffer: |
20 mM Hepes, 200 mM NaCl, 1 mM βME, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 30
|
A MademoiseLLE domain binding platform links the key RNA transporter to endosomes
PLOS Genetics 18(6):e1010269 (2022)
Devan S, Schott-Verdugo S, Müntjes K, Bismar L, Reiners J, Hachani E, Schmitt L, Höppner A, Smits S, Gohlke H, Feldbrügge M, Mitchell A
|
| RgGuinier |
5.6 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
UniProt ID: Q61771 (475-747) Kinesin-like protein KIF3B
UniProt ID: P70188 (1-693) Kinesin-associated protein 3
UniProt ID: P28741 (481-701) Kinesin-like protein KIF3A
|
|
|

|
| Sample: |
Kinesin-like protein KIF3B monomer, 32 kDa Mus musculus protein
Kinesin-associated protein 3 monomer, 81 kDa Mus musculus protein
Kinesin-like protein KIF3A monomer, 28 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Jun 24
|
The two-step cargo recognition mechanism of heterotrimeric kinesin.
EMBO Rep :e56864 (2023)
Jiang X, Ogawa T, Yonezawa K, Shimizu N, Ichinose S, Uchihashi T, Nagaike W, Moriya T, Adachi N, Kawasaki M, Dohmae N, Senda T, Hirokawa N
|
| RgGuinier |
5.8 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
557 |
nm3 |
|
|
UniProt ID: Q61315 (338-1010) Adenomatous polyposis coli protein (N-terminal ARM domain)
UniProt ID: P28741 (481-701) Kinesin-like protein KIF3A
UniProt ID: Q61771 (475-747) Kinesin-like protein KIF3B
UniProt ID: P70188 (1-692) Kinesin-associated protein 3
|
|
|

|
| Sample: |
Adenomatous polyposis coli protein (N-terminal ARM domain) monomer, 75 kDa Mus musculus protein
Kinesin-like protein KIF3A monomer, 28 kDa Mus musculus protein
Kinesin-like protein KIF3B monomer, 32 kDa Mus musculus protein
Kinesin-associated protein 3 monomer, 81 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 17
|
The two-step cargo recognition mechanism of heterotrimeric kinesin.
EMBO Rep :e56864 (2023)
Jiang X, Ogawa T, Yonezawa K, Shimizu N, Ichinose S, Uchihashi T, Nagaike W, Moriya T, Adachi N, Kawasaki M, Dohmae N, Senda T, Hirokawa N
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
763 |
nm3 |
|
|
UniProt ID: B6SVZ0 (69-567) Beta-amylase
|
|
|
|
| Sample: |
Beta-amylase tetramer, 231 kDa Zea mays protein
|
| Buffer: |
50 mM HEPES, 25 mM NaCl, and 0.2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 7
|
The BAM7
gene in Zea mays
encodes a protein with similar structural and catalytic properties to Arabidopsis
BAM2
Acta Crystallographica Section D Structural Biology 78(5) (2022)
Ravenburg C, Riney M, Monroe J, Berndsen C
|
| RgGuinier |
5.3 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
615 |
nm3 |
|
|
UniProt ID: B6SVZ0 (69-567) Beta-amylase
|
|
|
|
| Sample: |
Beta-amylase tetramer, 231 kDa Zea mays protein
|
| Buffer: |
50 mM HEPES, 25 mM NaCl, and 0.2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 7
|
The BAM7
gene in Zea mays
encodes a protein with similar structural and catalytic properties to Arabidopsis
BAM2
Acta Crystallographica Section D Structural Biology 78(5) (2022)
Ravenburg C, Riney M, Monroe J, Berndsen C
|
| RgGuinier |
5.1 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
467 |
nm3 |
|
|
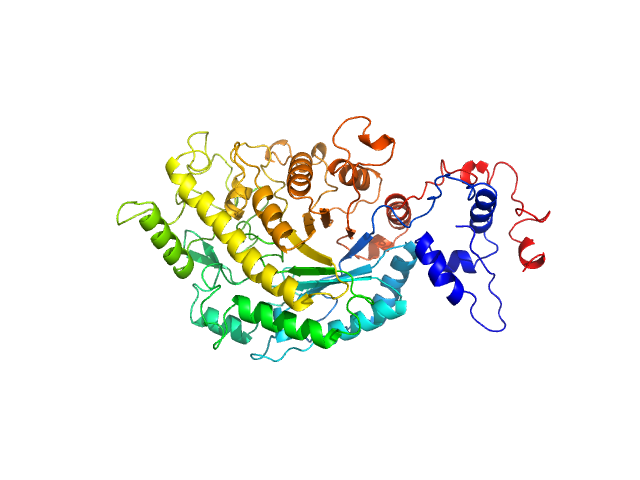
UniProt ID: Q9LIR6 (42-575) Beta-amylase 1, chloroplastic
|
|
|
|
| Sample: |
Beta-amylase 1, chloroplastic monomer, 65 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM MES, 100 mM NaCl, 1 mM DTT, pH: 6.7 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 22
|
The BAM7
gene in Zea mays
encodes a protein with similar structural and catalytic properties to Arabidopsis
BAM2
Acta Crystallographica Section D Structural Biology 78(5) (2022)
Ravenburg C, Riney M, Monroe J, Berndsen C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
88 |
nm3 |
|
|