UniProt ID: P0CG47 (1-76) Polyubiquitin-B
|
|
|
|
| Sample: |
Polyubiquitin-B monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02% NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Oct 8
|
Mechanistic insights into enhancement or inhibition of phase separation by different polyubiquitin chains.
EMBO Rep :e55056 (2022)
Dao TP, Yang Y, Presti MF, Cosgrove MS, Hopkins JB, Ma W, Loh SN, Castañeda CA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
UniProt ID: P0CG47 (1-76) Polyubiquitin-B
|
|
|
|
| Sample: |
Polyubiquitin-B monomer, 34 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02% NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Oct 8
|
Mechanistic insights into enhancement or inhibition of phase separation by different polyubiquitin chains.
EMBO Rep :e55056 (2022)
Dao TP, Yang Y, Presti MF, Cosgrove MS, Hopkins JB, Ma W, Loh SN, Castañeda CA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
UniProt ID: P0CG47 (1-76) Polyubiquitin-B
|
|
|
|
| Sample: |
Polyubiquitin-B tetramer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02% NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Oct 8
|
Mechanistic insights into enhancement or inhibition of phase separation by different polyubiquitin chains.
EMBO Rep :e55056 (2022)
Dao TP, Yang Y, Presti MF, Cosgrove MS, Hopkins JB, Ma W, Loh SN, Castañeda CA
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
UniProt ID: P9WNC1 (1-255) Endonuclease 8 2
UniProt ID: P9WGD5 (1-164) Single-stranded DNA-binding protein
|
|
|

|
| Sample: |
Endonuclease 8 2 dimer, 108 kDa Mycobacterium tuberculosis (strain … protein
Single-stranded DNA-binding protein octamer, 139 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Jul 10
|
Identification and characterization of MtbNei2-SSB forms novel pre-replicative BER complex in Mycobacterium tuberculosis.
Jyoti Vishwakarma
|
| RgGuinier |
5.5 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
734 |
nm3 |
|
|
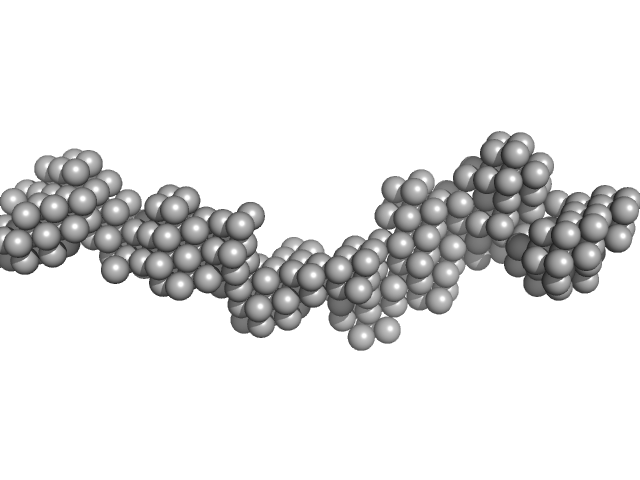
UniProt ID: Q15262 (28-752) Receptor-type tyrosine-protein phosphatase kappa
|
|
|
|
| Sample: |
Receptor-type tyrosine-protein phosphatase kappa monomer, 82 kDa Homo sapiens protein
|
| Buffer: |
50 mM MES, 250 mM NaCl, 3% v/v glycerol,, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Dec 13
|
Determinants of receptor tyrosine phosphatase homophilic adhesion: structural comparison of PTPRK and PTPRM extracellular domains
Journal of Biological Chemistry :102750 (2022)
Hay I, Shamin M, Caroe E, Mohammed A, Svergun D, Jeffries C, Graham S, Sharpe H, Deane J
|
| RgGuinier |
7.0 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
252 |
nm3 |
|
|
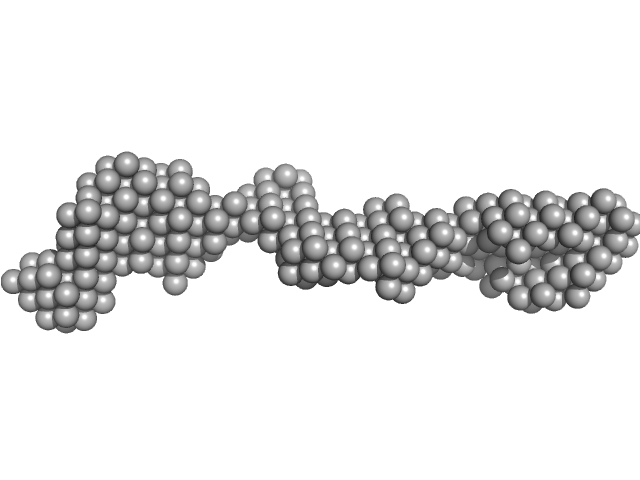
UniProt ID: P28827 (19-742) Receptor-type tyrosine-protein phosphatase mu
|
|
|
|
| Sample: |
Receptor-type tyrosine-protein phosphatase mu monomer, 82 kDa Homo sapiens protein
|
| Buffer: |
50 mM MES, 250 mM NaCl, 3% v/v glycerol,, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Dec 13
|
Determinants of receptor tyrosine phosphatase homophilic adhesion: structural comparison of PTPRK and PTPRM extracellular domains
Journal of Biological Chemistry :102750 (2022)
Hay I, Shamin M, Caroe E, Mohammed A, Svergun D, Jeffries C, Graham S, Sharpe H, Deane J
|
| RgGuinier |
7.2 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
255 |
nm3 |
|
|
UniProt ID: P02769 (25-607) Bovine Serum Albumin
|
|
|
|
| Sample: |
Bovine Serum Albumin trimer, 199 kDa Bos taurus protein
|
| Buffer: |
20 mM Hepes, 150mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Sep 24
|
SEC-SAXS: Experimental set-up and software developments build up a powerful tool.
Methods Enzymol 677:221-249 (2022)
Pérez J, Thureau A, Vachette P
|
| RgGuinier |
5.2 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
370 |
nm3 |
|
|
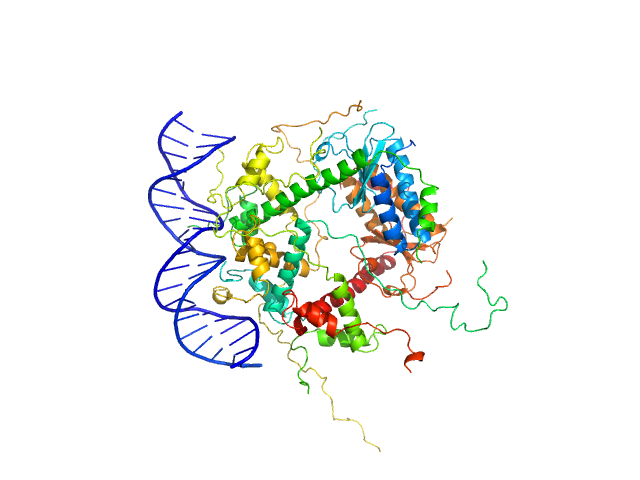
UniProt ID: P58093 (1-80) Antitoxin ParD
UniProt ID: Q9KMJ0 (1-99) Toxin
UniProt ID: None (None-None) 21-bp DNA operator fragment
|
|
|
|
| Sample: |
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
21-bp DNA operator fragment monomer, 13 kDa Vibrio cholerae O1 DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jul 18
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
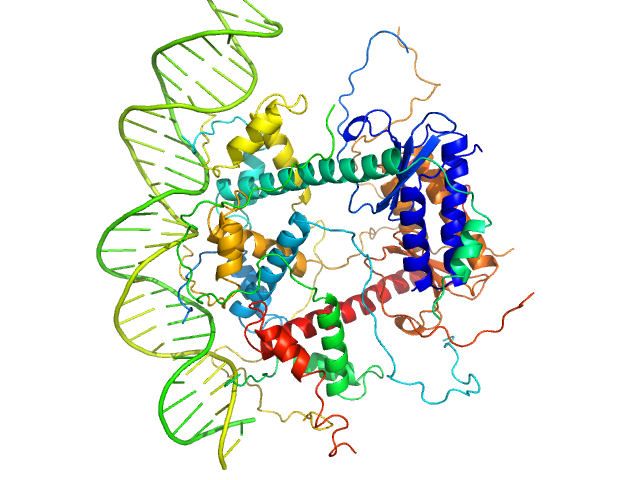
UniProt ID: P58093 (1-80) Antitoxin ParD
UniProt ID: Q9KMJ0 (1-99) Toxin
UniProt ID: None (None-None) 31-bp DNA operator box
|
|
|
|
| Sample: |
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
31-bp DNA operator box monomer, 19 kDa Vibrio cholerae O1 DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 4
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
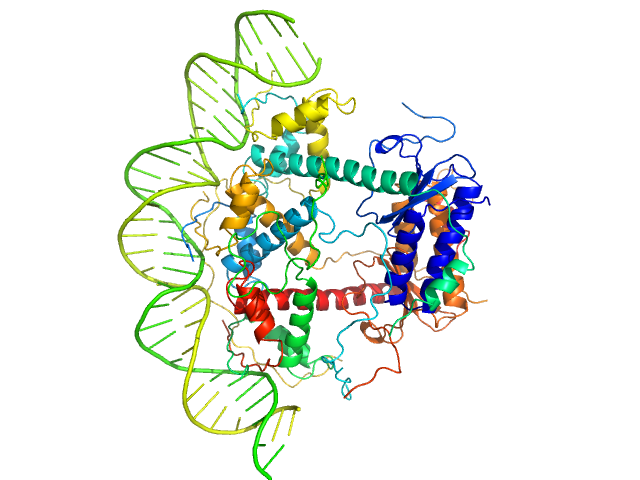
UniProt ID: P58093 (1-80) Antitoxin ParD
UniProt ID: Q9KMJ0 (1-99) Toxin
UniProt ID: None (None-None) 33-bp DNA operator fragment
|
|
|
|
| Sample: |
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
33-bp DNA operator fragment monomer, 20 kDa Vibrio cholerae O1 DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jul 18
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
150 |
nm3 |
|
|