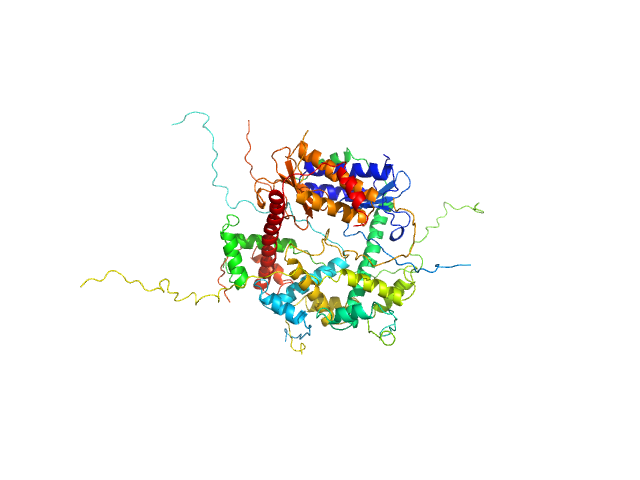
UniProt ID: P58093 (1-80) Antitoxin ParD
UniProt ID: Q9KMJ0 (1-99) Toxin
|
|
|
|
| Sample: |
Antitoxin ParD hexamer, 54 kDa Vibrio cholerae serotype … protein
Toxin, 25 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Mar 6
|
Toxin:antitoxin ratio sensing autoregulation of the Vibrio cholerae parDE2 module.
Sci Adv 10(1):eadj2403 (2024)
Garcia-Rodriguez G, Girardin Y, Kumar Singh R, Volkov AN, Van Dyck J, Muruganandam G, Sobott F, Charlier D, Loris R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
UniProt ID: Q03164 (3754-3969) Histone-lysine N-methyltransferase 2A
UniProt ID: P61964 (1-334) WD repeat-containing protein 5
UniProt ID: Q15291 (1-538) Retinoblastoma-binding protein 5
UniProt ID: Q9UBL3-3 (95-534) Set1/Ash2 histone methyltransferase complex subunit ASH2
|
|
|
|
| Sample: |
Histone-lysine N-methyltransferase 2A monomer, 25 kDa Homo sapiens protein
WD repeat-containing protein 5 monomer, 37 kDa Homo sapiens protein
Retinoblastoma-binding protein 5 monomer, 59 kDa Homo sapiens protein
Set1/Ash2 histone methyltransferase complex subunit ASH2 monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM TECP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Feb 1
|
DPY30 acts as an ASH2L-specific stabilizer to stimulate the enzyme activity of MLL family methyltransferases on different substrates.
iScience 25(9):104948 (2022)
Zhao L, Huang N, Mencius J, Li Y, Xu Y, Zheng Y, He W, Li N, Zheng J, Zhuang M, Quan S, Chen Y
|
| RgGuinier |
5.2 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
386 |
nm3 |
|
|
UniProt ID: Q03164 (3754-3969) Histone-lysine N-methyltransferase 2A
UniProt ID: P61964 (1-334) WD repeat-containing protein 5
UniProt ID: Q15291 (1-538) Retinoblastoma-binding protein 5
UniProt ID: Q9UBL3-3 (95-534) Set1/Ash2 histone methyltransferase complex subunit ASH2
UniProt ID: Q9C005 (1-99) Protein dpy-30 homolog
|
|
|
|
| Sample: |
Histone-lysine N-methyltransferase 2A monomer, 25 kDa Homo sapiens protein
WD repeat-containing protein 5 monomer, 37 kDa Homo sapiens protein
Retinoblastoma-binding protein 5 monomer, 59 kDa Homo sapiens protein
Set1/Ash2 histone methyltransferase complex subunit ASH2 monomer, 60 kDa Homo sapiens protein
Protein dpy-30 homolog dimer, 23 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM TECP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Feb 1
|
DPY30 acts as an ASH2L-specific stabilizer to stimulate the enzyme activity of MLL family methyltransferases on different substrates.
iScience 25(9):104948 (2022)
Zhao L, Huang N, Mencius J, Li Y, Xu Y, Zheng Y, He W, Li N, Zheng J, Zhuang M, Quan S, Chen Y
|
| RgGuinier |
5.4 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
411 |
nm3 |
|
|
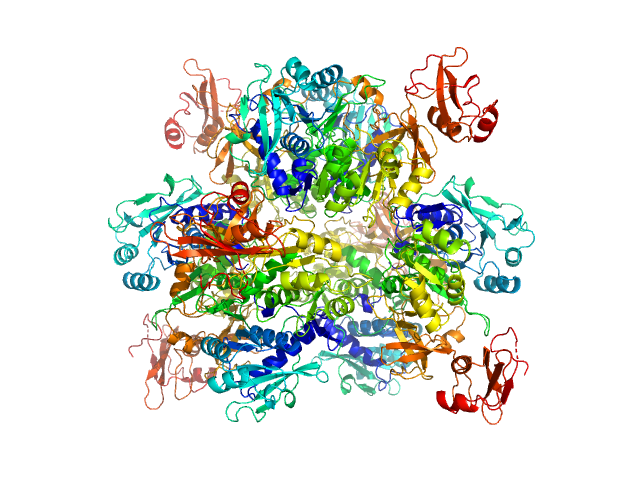
UniProt ID: P38137 (1-543) Oxalate--CoA ligase
|
|
|
|
| Sample: |
Oxalate--CoA ligase, 363 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol Chem (2023)
Bürgi J, Lill P, Giannopoulou EA, Jeffries CM, Chojnowski G, Raunser S, Gatsogiannis C, Wilmanns M
|
| RgGuinier |
4.7 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
565 |
nm3 |
|
|
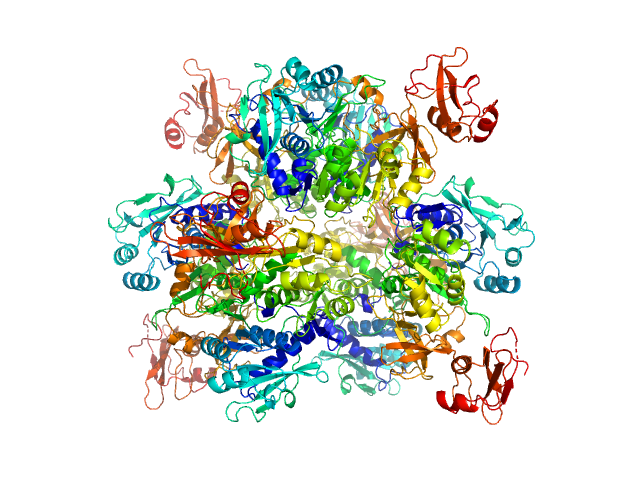
UniProt ID: P38137 (1-543) Oxalate--CoA ligase
|
|
|
|
| Sample: |
Oxalate--CoA ligase, 363 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol Chem (2023)
Bürgi J, Lill P, Giannopoulou EA, Jeffries CM, Chojnowski G, Raunser S, Gatsogiannis C, Wilmanns M
|
| RgGuinier |
4.5 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
432 |
nm3 |
|
|
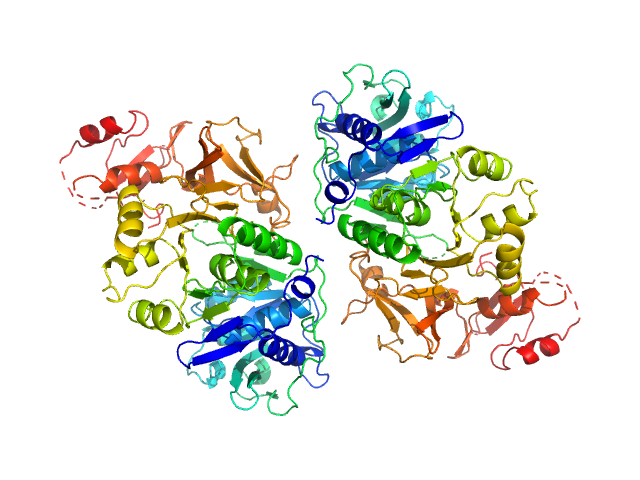
UniProt ID: P38137 (1-543) Oxalate--CoA ligase (K352D)
|
|
|
|
| Sample: |
Oxalate--CoA ligase (K352D) dimer, 121 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol Chem (2023)
Bürgi J, Lill P, Giannopoulou EA, Jeffries CM, Chojnowski G, Raunser S, Gatsogiannis C, Wilmanns M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
163 |
nm3 |
|
|
UniProt ID: P38137 (1-543) Oxalate--CoA ligase (K352D)
|
|
|
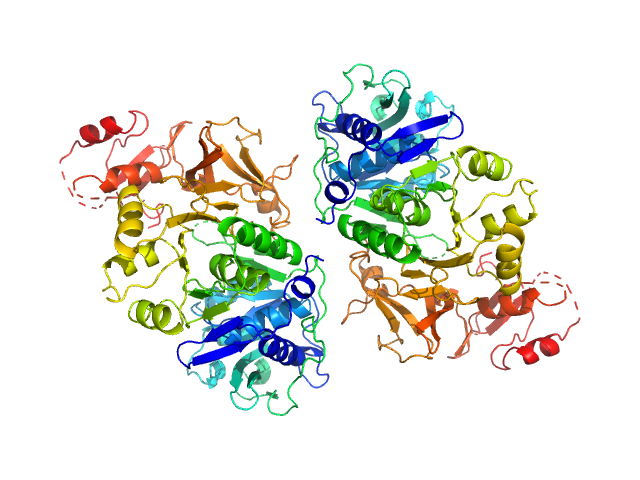
|
| Sample: |
Oxalate--CoA ligase (K352D) dimer, 121 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
50 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 18
|
Asymmetric horseshoe-like assembly of peroxisomal yeast oxalyl-CoA synthetase.
Biol Chem (2023)
Bürgi J, Lill P, Giannopoulou EA, Jeffries CM, Chojnowski G, Raunser S, Gatsogiannis C, Wilmanns M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
UniProt ID: P23025 (1-239) DNA repair protein complementing XP-A cells
UniProt ID: P27694 (185-616) Replication protein A 70 kDa DNA-binding subunit
UniProt ID: P15927 (45-270) Replication protein A 32 kDa subunit
UniProt ID: P35244 (1-121) Replication protein A 14 kDa subunit
UniProt ID: None (None-None) 3-prime ss-ds DNA junction NER model substrate
|
|
|
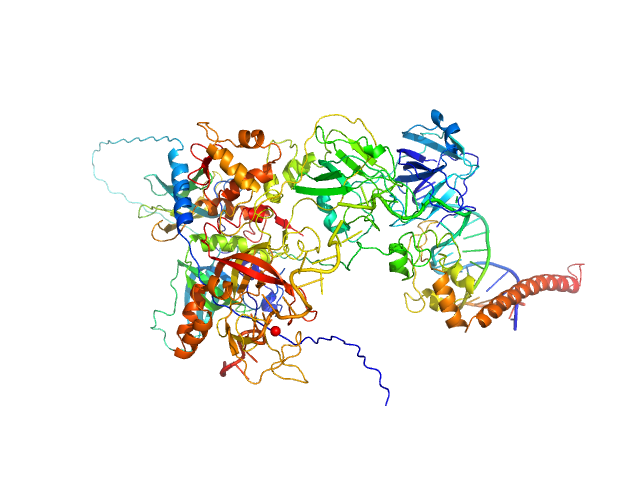
|
| Sample: |
DNA repair protein complementing XP-A cells monomer, 27 kDa Homo sapiens protein
Replication protein A 70 kDa DNA-binding subunit monomer, 49 kDa Homo sapiens protein
Replication protein A 32 kDa subunit monomer, 25 kDa Homo sapiens protein
Replication protein A 14 kDa subunit monomer, 14 kDa Homo sapiens protein
3-prime ss-ds DNA junction NER model substrate monomer, 17 kDa DNA
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 2% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Mar 4
|
Two interaction surfaces between XPA and RPA organize the preincision complex in nucleotide excision repair
Proceedings of the National Academy of Sciences 119(34) (2022)
Kim M, Kim H, D’Souza A, Gallagher K, Jeong E, Topolska-Wós A, Ogorodnik Le Meur K, Tsai C, Tsai M, Kee M, Tainer J, Yeo J, Chazin W, Schärer O
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
UniProt ID: P35244 (1-121) Replication protein A 14 kDa subunit
UniProt ID: P23025 (1-239) DNA repair protein complementing XP-A cells
UniProt ID: P27694 (185-616) Replication protein A 70 kDa DNA-binding subunit
UniProt ID: P15927 (45-270) Replication protein A 32 kDa subunit
UniProt ID: None (None-None) 5-prime ss-ds DNA junction NER model substrate
|
|
|
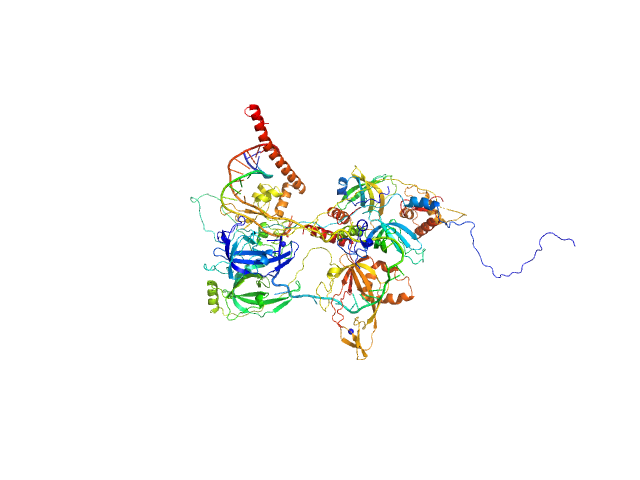
|
| Sample: |
Replication protein A 14 kDa subunit monomer, 14 kDa Homo sapiens protein
DNA repair protein complementing XP-A cells monomer, 27 kDa Homo sapiens protein
Replication protein A 70 kDa DNA-binding subunit monomer, 49 kDa Homo sapiens protein
Replication protein A 32 kDa subunit monomer, 25 kDa Homo sapiens protein
5-prime ss-ds DNA junction NER model substrate monomer, 17 kDa DNA
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 2% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Mar 4
|
Two interaction surfaces between XPA and RPA organize the preincision complex in nucleotide excision repair
Proceedings of the National Academy of Sciences 119(34) (2022)
Kim M, Kim H, D’Souza A, Gallagher K, Jeong E, Topolska-Wós A, Ogorodnik Le Meur K, Tsai C, Tsai M, Kee M, Tainer J, Yeo J, Chazin W, Schärer O
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
UniProt ID: P51668 (1-147) Ubiquitin-conjugating enzyme E2 D1 (S22R, C85K, D87S)
UniProt ID: P0CG48 (1-76) Ubiquitin
|
|
|
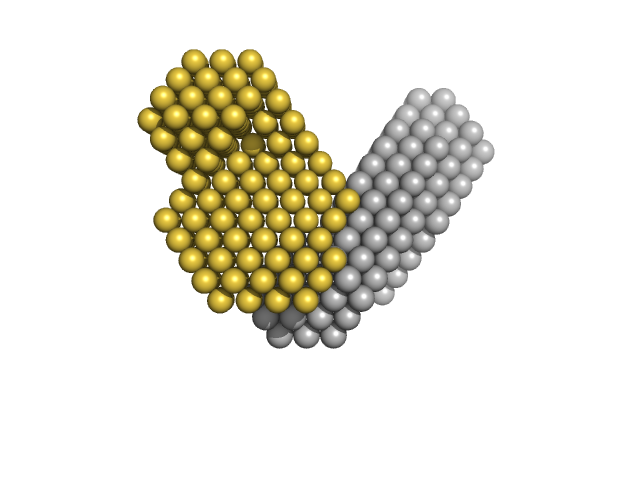
|
| Sample: |
Ubiquitin-conjugating enzyme E2 D1 (S22R, C85K, D87S) monomer, 17 kDa Homo sapiens protein
Ubiquitin monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SANS
data collected at Quokka - Small Angle Neutron Scattering, Australian Centre for Neutron Scattering (ANSTO) on 2019 May 24
|
Production and characterisation of modularly deuterated UBE2D1–Ub conjugate by small angle neutron and X-ray scattering
European Biophysics Journal (2022)
Pietras Z, Duff A, Morad V, Wood K, Jeffries C, Sunnerhagen M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.4 |
nm |
|
|