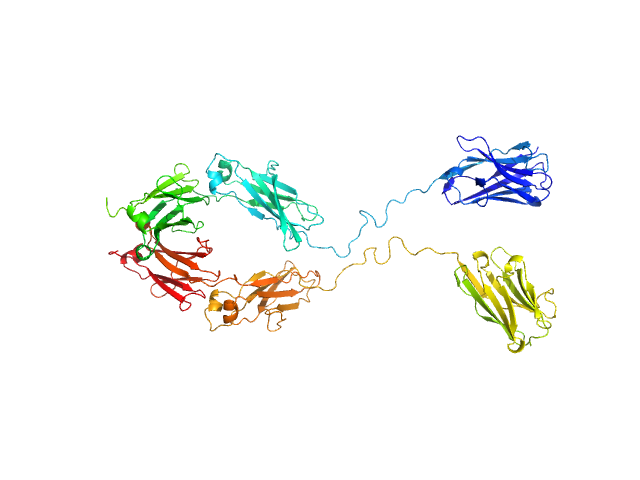
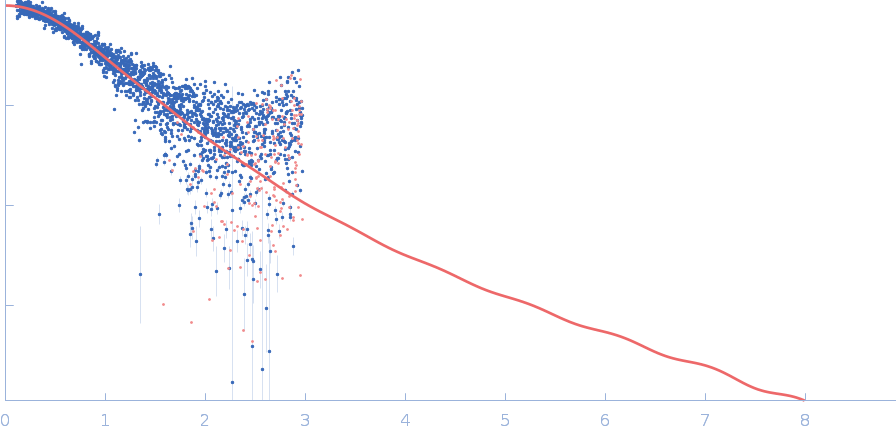
UniProt ID: None (None-None) Immunoglobulin heavy constant gamma 1
|
|
|
|
| Sample: |
Immunoglobulin heavy constant gamma 1 monomer, 78 kDa Homo sapiens protein
|
| Buffer: |
20 mM L-histidine, 138 mM NaCl, 2.6 mM KCl buffer, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Sep 28
|
The solution structure of the heavy chain-only C5-Fc nanobody reveals exposed variable regions that are optimal for COVID-19 antigen interactions.
J Biol Chem :105337 (2023)
Gao X, Thrush JW, Gor J, Naismith JH, Owens RJ, Perkins SJ
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
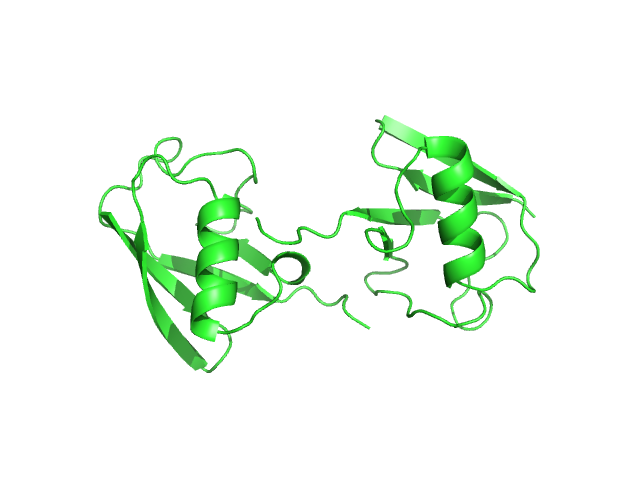
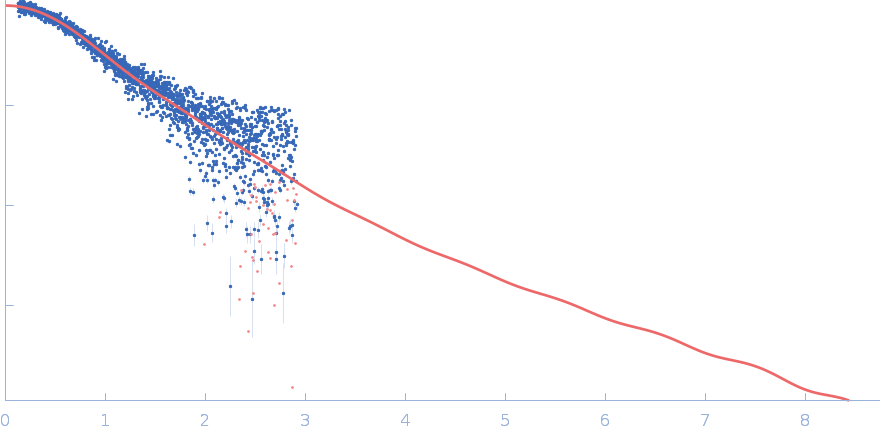
UniProt ID: P0CG48 (1-152) Polyubiquitin-C
|
|
|
|
| Sample: |
Polyubiquitin-C monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 1
|
Stretching the chains: the destabilizing impact of Cu(2+) and Zn(2+) ions on K48-linked diubiquitin.
Dalton Trans (2023)
Mangini V, Grasso G, Belviso BD, Sciacca MFM, Lanza V, Caliandro R, Milardi D
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
UniProt ID: P0CG48 (1-152) Polyubiquitin-C
|
|
|
|
| Sample: |
Polyubiquitin-C monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 1
|
Stretching the chains: the destabilizing impact of Cu(2+) and Zn(2+) ions on K48-linked diubiquitin.
Dalton Trans (2023)
Mangini V, Grasso G, Belviso BD, Sciacca MFM, Lanza V, Caliandro R, Milardi D
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
UniProt ID: P0CG48 (1-152) Polyubiquitin-C
|
|
|
|
| Sample: |
Polyubiquitin-C monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 1
|
Stretching the chains: the destabilizing impact of Cu(2+) and Zn(2+) ions on K48-linked diubiquitin.
Dalton Trans (2023)
Mangini V, Grasso G, Belviso BD, Sciacca MFM, Lanza V, Caliandro R, Milardi D
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
UniProt ID: Q5HGG6 (2-155) Ribosome maturation factor RimP
UniProt ID: Q5HID0 (2-136) 30S ribosomal protein S12
|
|
|

|
| Sample: |
Ribosome maturation factor RimP monomer, 18 kDa Staphylococcus aureus (strain … protein
30S ribosomal protein S12 monomer, 15 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
50 mM sodium phosphate, 200 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at Xeuss 3.0 SAXS/WAXS System, JINR on 2023 Feb 16
|
Structural aspects of RimP binding on small ribosomal subunit from Staphylococcus aureus.
Structure (2023)
Garaeva N, Fatkhullin B, Murzakhanov F, Gafurov M, Golubev A, Bikmullin A, Glazyrin M, Kieffer B, Jenner L, Klochkov V, Aganov A, Rogachev A, Ivankov O, Validov S, Yusupov M, Usachev K
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
UniProt ID: P03372 (1-184) Estrogen receptor
|
|
|
|
| Sample: |
Estrogen receptor monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 150 mM NaCl, 0.5 mM EDTA, 0.1 mM PMSF, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jul 21
|
The sequence–structure–function relationship of intrinsic ERα disorder
Nature (2025)
Du Z, Wang H, Luo S, Yun Z, Wu C, Yang W, Buck M, Zheng W, Hansen A, Kao H, Yang S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
UniProt ID: Q12904 (147-312) Aminoacyl tRNA synthase complex-interacting multifunctional protein 1
|
|
|
|
| Sample: |
Aminoacyl tRNA synthase complex-interacting multifunctional protein 1 monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2022 Oct 12
|
Solution structure of the EMAPII domain of human aminoacyl-tRNA synthetase complex
Michal Taube
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
UniProt ID: A0A178VHQ2 (54-131) CP12-2 (D77G)
|
|
|
|
| Sample: |
CP12-2 (D77G) monomer, 8 kDa Arabidopsis thaliana protein
|
| Buffer: |
25 mM potassium phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 15
|
Conformational Disorder Analysis of the Conditionally Disordered Protein CP12 from Arabidopsis thaliana in Its Different Redox States
International Journal of Molecular Sciences 24(11):9308 (2023)
Del Giudice A, Gurrieri L, Galantini L, Fanti S, Trost P, Sparla F, Fermani S
|
| RgGuinier |
2.0 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
UniProt ID: A0A178VHQ2 (54-131) CP12-2 (D77G)
|
|
|
|
| Sample: |
CP12-2 (D77G) monomer, 8 kDa Arabidopsis thaliana protein
|
| Buffer: |
25 mM potassium phosphate, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 15
|
Conformational Disorder Analysis of the Conditionally Disordered Protein CP12 from Arabidopsis thaliana in Its Different Redox States
International Journal of Molecular Sciences 24(11):9308 (2023)
Del Giudice A, Gurrieri L, Galantini L, Fanti S, Trost P, Sparla F, Fermani S
|
| RgGuinier |
2.3 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
UniProt ID: D8SB57 (10-76) C3H1-type domain-containing protein
|
|
|
|
| Sample: |
C3H1-type domain-containing protein monomer, 8 kDa Selaginella moellendorffii protein
|
| Buffer: |
20 mM sodium phosphate, 100 mM KCl, 25 µM ZnSO4, 2 mM β-mercaptoethanol, 5 % glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Nov 9
|
Solution scattering of the tandem zinc finger (TZF) domain from the tristetraprolin (TTP) family member found in Selaginella moellendorffii (spikemoss) in the presence and absence of 9-mer RNA
Stephanie Hicks
|
| RgGuinier |
1.8 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
15 |
nm3 |
|
|