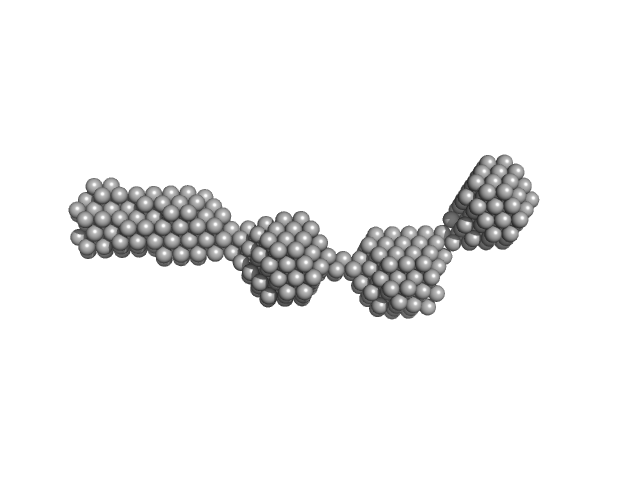
UniProt ID: P11709 (2-440) Nuclear fusion protein BIK1
|
|
|
|
| Sample: |
Nuclear fusion protein BIK1 dimer, 106 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM Tris-HCl pH 7.5, 500 mM NaCl, 2% glycerol, 1 mM DTT, pH: |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Jul 13
|
Phase separation of a microtubule plus-end tracking protein into a fluid fractal network
(2024)
Czub M, Uliana F, Grubić T, Padeste C, Rosowski K, Dufresne E, Menzel A, Vakonakis I, Gasser U, Steinmetz M
|
| RgGuinier |
8.8 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
467 |
nm3 |
|
|
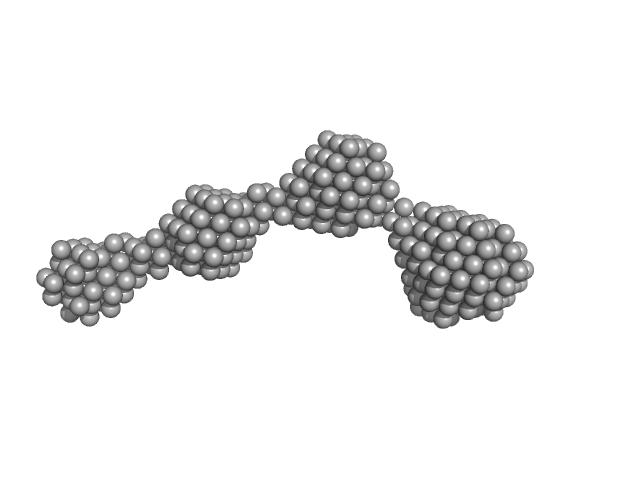
UniProt ID: P11709 (182-389) Nuclear fusion protein BIK1
|
|
|
|
| Sample: |
Nuclear fusion protein BIK1 dimer, 55 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM Tris-HCl pH 7.5, 500 mM NaCl, 2% glycerol, 1 mM DTT, pH: |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Jul 13
|
Phase separation of a microtubule plus-end tracking protein into a fluid fractal network
(2024)
Czub M, Uliana F, Grubić T, Padeste C, Rosowski K, Dufresne E, Menzel A, Vakonakis I, Gasser U, Steinmetz M
|
| RgGuinier |
8.0 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
282 |
nm3 |
|
|
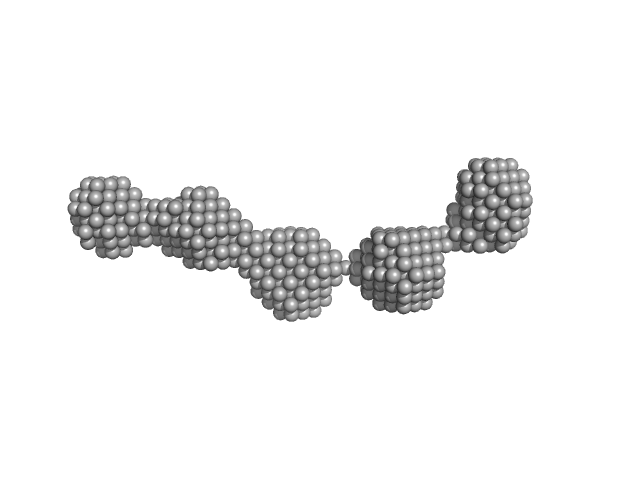
UniProt ID: P11709 (2-436) Nuclear fusion protein BIK1
|
|
|
|
| Sample: |
Nuclear fusion protein BIK1 dimer, 105 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM Tris-HCl pH 7.5, 500 mM NaCl, 2% glycerol, 1 mM DTT, pH: |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Jul 13
|
Phase separation of a microtubule plus-end tracking protein into a fluid fractal network
(2024)
Czub M, Uliana F, Grubić T, Padeste C, Rosowski K, Dufresne E, Menzel A, Vakonakis I, Gasser U, Steinmetz M
|
| RgGuinier |
9.4 |
nm |
| Dmax |
29.4 |
nm |
| VolumePorod |
632 |
nm3 |
|
|
UniProt ID: A0A3P7XL18 (63-297) Alarmin release inhibitor (Δ1-62)
|
|
|

|
| Sample: |
Alarmin release inhibitor (Δ1-62) monomer, 26 kDa Heligmosomoides polygyrus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, 5% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Feb 9
|
Structural basis for IL-33 recognition and its antagonism by the helminth effector protein HpARI2.
Nat Commun 15(1):5226 (2024)
Jamwal A, Colomb F, McSorley HJ, Higgins MK
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
UniProt ID: A0A3P7XL18 (126-297) Alarmin release inhibitor (Δ1-125; N175Q, N190Q)
|
|
|

|
| Sample: |
Alarmin release inhibitor (Δ1-125; N175Q, N190Q) monomer, 19 kDa Heligmosomoides polygyrus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, 5% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Feb 9
|
Structural basis for IL-33 recognition and its antagonism by the helminth effector protein HpARI2.
Nat Commun 15(1):5226 (2024)
Jamwal A, Colomb F, McSorley HJ, Higgins MK
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
UniProt ID: A0A3P7XL18 (63-297) Alarmin release inhibitor (Δ1-62)
UniProt ID: Q8BVZ5 (109-266) Interleukin-33 (L179V)
|
|
|

|
| Sample: |
Alarmin release inhibitor (Δ1-62) monomer, 26 kDa Heligmosomoides polygyrus protein
Interleukin-33 (L179V) monomer, 18 kDa Mus musculus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, 5% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Feb 9
|
Structural basis for IL-33 recognition and its antagonism by the helminth effector protein HpARI2.
Nat Commun 15(1):5226 (2024)
Jamwal A, Colomb F, McSorley HJ, Higgins MK
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
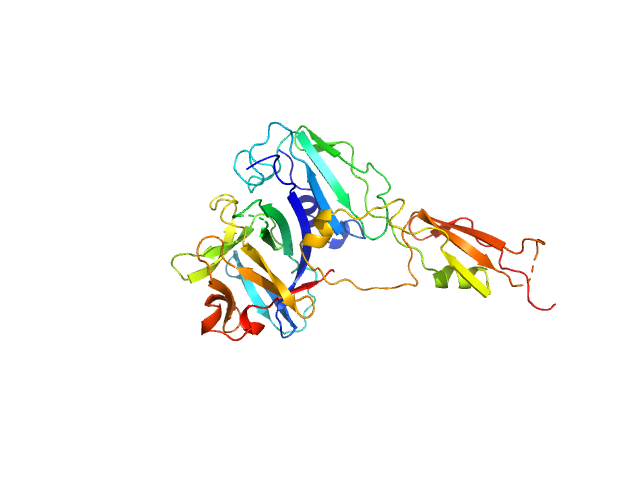
UniProt ID: Q8BVZ5 (109-266) Interleukin-33 (L179V)
UniProt ID: A0A3P7XL18 (126-297) Alarmin release inhibitor (Δ1-125; N175Q, N190Q)
|
|
|
|
| Sample: |
Interleukin-33 (L179V) monomer, 18 kDa Mus musculus protein
Alarmin release inhibitor (Δ1-125; N175Q, N190Q) monomer, 19 kDa Heligmosomoides polygyrus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, 5% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Feb 9
|
Structural basis for IL-33 recognition and its antagonism by the helminth effector protein HpARI2.
Nat Commun 15(1):5226 (2024)
Jamwal A, Colomb F, McSorley HJ, Higgins MK
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
UniProt ID: F4IED2 (1-274) NAC domain-containing protein 13
|
|
|
|
| Sample: |
NAC domain-containing protein 13 dimer, 63 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM NaNO3, 5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Oct 3
|
Allovalent scavenging of activation domains in the transcription factor ANAC013 gears transcriptional regulation.
Nucleic Acids Res 53(4) (2025)
Delaforge E, Due AD, Theisen FF, Morffy N, O'Shea C, Blackledge M, Strader LC, Skriver K, Kragelund BB
|
| RgGuinier |
3.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
118 |
nm3 |
|
|
UniProt ID: F4IED2 (161-274) NAC domain-containing protein 13
|
|
|
|
| Sample: |
NAC domain-containing protein 13 monomer, 13 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM NaNO3, 5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Oct 3
|
Allovalent scavenging of activation domains in the transcription factor ANAC013 gears transcriptional regulation.
Nucleic Acids Res 53(4) (2025)
Delaforge E, Due AD, Theisen FF, Morffy N, O'Shea C, Blackledge M, Strader LC, Skriver K, Kragelund BB
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
UniProt ID: Q9H9Q4 (1-299) Non-homologous end-joining factor 1
|
|
|
|
| Sample: |
Non-homologous end-joining factor 1 dimer, 67 kDa Homo sapiens protein
|
| Buffer: |
20 mM Bis-tris, 150 mM KCl, 1 mM EDTA, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Mar 13
|
Multivalent interactions of the disordered regions of XLF and XRCC4 foster robust cellular NHEJ and drive the formation of ligation-boosting condensates in vitro.
Nat Struct Mol Biol (2024)
Vu DD, Bonucci A, Brenière M, Cisneros-Aguirre M, Pelupessy P, Wang Z, Carlier L, Bouvignies G, Cortes P, Aggarwal AK, Blackledge M, Gueroui Z, Belle V, Stark JM, Modesti M, Ferrage F
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
123 |
nm3 |
|
|