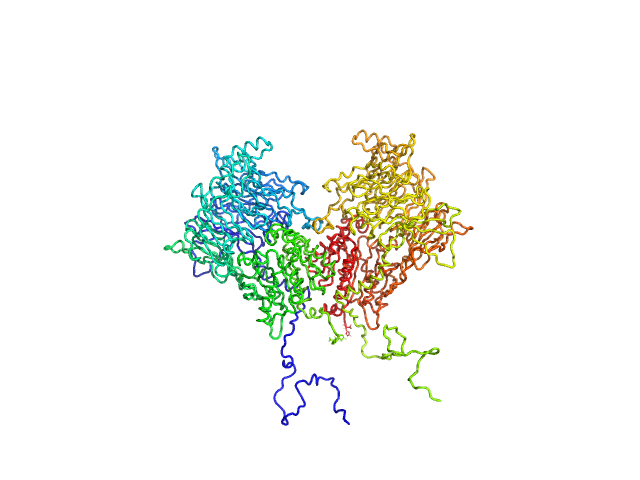
UniProt ID: Q6V1X1-1 (None-None) Isoform 1 of Dipeptidyl peptidase 8
|
|
|
|
| Sample: |
Isoform 1 of Dipeptidyl peptidase 8 dimer, 208 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Oct 14
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
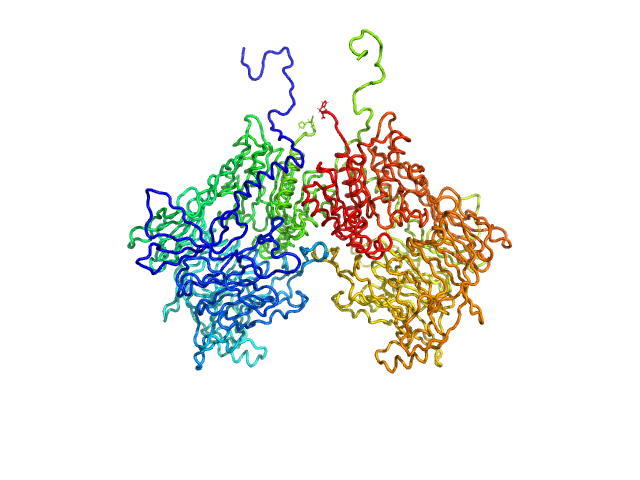
UniProt ID: Q86TI2-1 (1-863) Isoform 1 of Dipeptidyl peptidase 9
|
|
|
|
| Sample: |
Isoform 1 of Dipeptidyl peptidase 9 dimer, 198 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Sep 23
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
290 |
nm3 |
|
|
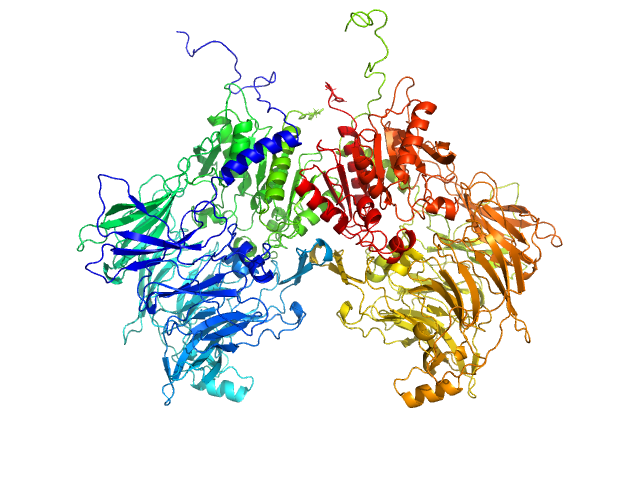
UniProt ID: Q86TI2-1 (1-863) Isoform 1 of Dipeptidyl peptidase 9
|
|
|
|
| Sample: |
Isoform 1 of Dipeptidyl peptidase 9 dimer, 198 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Sep 23
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.1 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
274 |
nm3 |
|
|
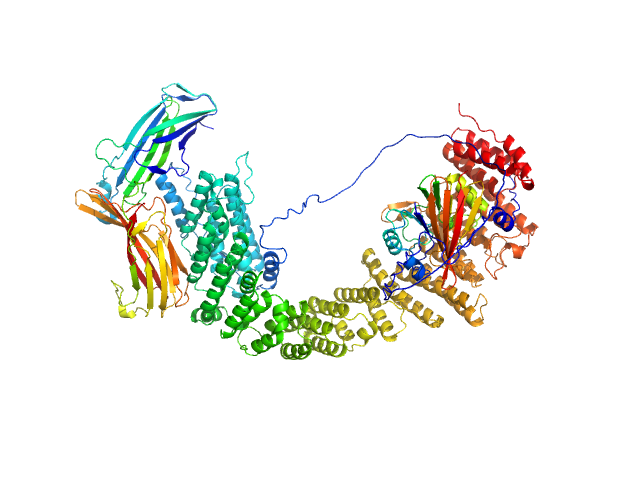
UniProt ID: Q7Z3J2 (1-963) VPS35 endosomal protein-sorting factor-like
UniProt ID: Q9UBQ0 (1-182) Vacuolar protein sorting-associated protein 29
UniProt ID: O14972 (1-297) Vacuolar protein sorting-associated protein 26C
|
|
|
|
| Sample: |
VPS35 endosomal protein-sorting factor-like monomer, 110 kDa Homo sapiens protein
Vacuolar protein sorting-associated protein 29 monomer, 21 kDa Homo sapiens protein
Vacuolar protein sorting-associated protein 26C monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 200 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Apr 28
|
Selective cargo and membrane recognition by SNX17 regulates its interaction with Retriever
EMBO Reports (2024)
Martín-González A, Méndez-Guzmán I, Zabala-Zearreta M, Quintanilla A, García-López A, Martínez-Lombardía E, Albesa-Jové D, Acosta J, Lucas M
|
| RgGuinier |
5.2 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
256 |
nm3 |
|
|
UniProt ID: O14972 (1-297) Vacuolar protein sorting-associated protein 26C
|
|
|
|
| Sample: |
Vacuolar protein sorting-associated protein 26C monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP,, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Apr 22
|
Selective cargo and membrane recognition by SNX17 regulates its interaction with Retriever
EMBO Reports (2024)
Martín-González A, Méndez-Guzmán I, Zabala-Zearreta M, Quintanilla A, García-López A, Martínez-Lombardía E, Albesa-Jové D, Acosta J, Lucas M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
UniProt ID: Q16643 (173-238) Drebrin
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDVV6_fit1_model1.png)
|
| Sample: |
Drebrin monomer, 10 kDa Homo sapiens protein
|
| Buffer: |
17 mM NaH2PO4, 3 mM Na2HPO4, 50 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jul 7
|
Dynamic Interchange of Local Residue-Residue Interactions in the Largely Extended Single Alpha-Helix in Drebrin
Biochemical Journal (2025)
Varga S, Péterfia B, Dudola D, Farkas V, Jeffries C, Permi P, Gáspári Z
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
UniProt ID: B1AKI9 (263-464) Isthmin-1 AMOP-domain
|
|
|
|
| Sample: |
Isthmin-1 AMOP-domain monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Pennsylvania State University on 2023 Jun 21
|
Crystal structure of Isthmin-1 and reassessment of its functional role in pre-adipocyte signaling.
Nat Commun 16(1):3580 (2025)
Li T, Stayrook SE, Li W, Wang Y, Li H, Zhang J, Liu Y, Klein DE
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
45264 |
nm3 |
|
|
UniProt ID: B1AKI9 (263-464) L-AMOP-IFEE
|
|
|
|
| Sample: |
L-AMOP-IFEE monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Pennsylvania State University on 2023 Jun 21
|
Crystal structure of Isthmin-1 and reassessment of its functional role in pre-adipocyte signaling.
Nat Commun 16(1):3580 (2025)
Li T, Stayrook SE, Li W, Wang Y, Li H, Zhang J, Liu Y, Klein DE
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
46874 |
nm3 |
|
|
UniProt ID: P23246 (214-707) Splicing factor, proline- and glutamine-rich
|
|
|
|
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 153 kDa Homo sapiens protein
|
| Buffer: |
500mM KNO3, 20mM HEPES, 5% glycerol, 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2022 Oct 22
|
Structural dynamics of IDR interactions in human SFPQ and implications for liquid–liquid phase separation
Acta Crystallographica Section D Structural Biology 81(7):357-379 (2025)
Koning H, Lai V, Sethi A, Chakraborty S, Ang C, Fox A, Duff A, Whitten A, Marshall A, Bond C
|
| RgGuinier |
7.9 |
nm |
| Dmax |
42.5 |
nm |
| VolumePorod |
485 |
nm3 |
|
|
UniProt ID: P23246 (1-598) Splicing Factor Proline/Glutamine rich (1-598)
|
|
|
|
| Sample: |
Splicing Factor Proline/Glutamine rich (1-598) dimer, 130 kDa Homo sapiens protein
|
| Buffer: |
500mM KNO3, 20mM HEPES, 5% glycerol, 1mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2023 Mar 13
|
Structural dynamics of IDR interactions in human SFPQ and implications for liquid–liquid phase separation
Acta Crystallographica Section D Structural Biology 81(7):357-379 (2025)
Koning H, Lai V, Sethi A, Chakraborty S, Ang C, Fox A, Duff A, Whitten A, Marshall A, Bond C
|
| RgGuinier |
7.6 |
nm |
| Dmax |
34.3 |
nm |
| VolumePorod |
400 |
nm3 |
|
|