UniProt ID: P23246 (1-598) Splicing Factor Proline/Glutamine rich (1-598)
|
|
|
|
| Sample: |
Splicing Factor Proline/Glutamine rich (1-598) dimer, 130 kDa Homo sapiens protein
|
| Buffer: |
150 mM KCl, 20 mM HEPES, 5% glycerol, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2023 Mar 13
|
Structural dynamics of IDR interactions in human SFPQ and implications for liquid–liquid phase separation
Acta Crystallographica Section D Structural Biology 81(7):357-379 (2025)
Koning H, Lai V, Sethi A, Chakraborty S, Ang C, Fox A, Duff A, Whitten A, Marshall A, Bond C
|
| RgGuinier |
6.1 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
315 |
nm3 |
|
|
UniProt ID: P04968 (2-514) L-threonine dehydratase biosynthetic IlvA
|
|
|
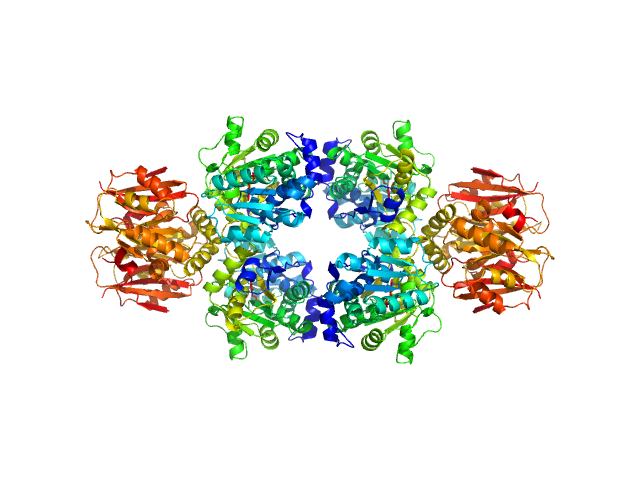
|
| Sample: |
L-threonine dehydratase biosynthetic IlvA tetramer, 233 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM KPO4, 200 mM NaCl, 0.1 mM TCEP,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 14
|
Isoleucine Binding and Regulation of Escherichia coli and Staphylococcus aureus Threonine Dehydratase (IlvA).
Biochemistry (2025)
Yun MK, Subramanian C, Miller K, Jackson P, Radka CD, Rock CO
|
| RgGuinier |
4.5 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
284 |
nm3 |
|
|
UniProt ID: P04968 (335-514) L-threonine dehydratase biosynthetic IlvA Regulatory domain
|
|
|
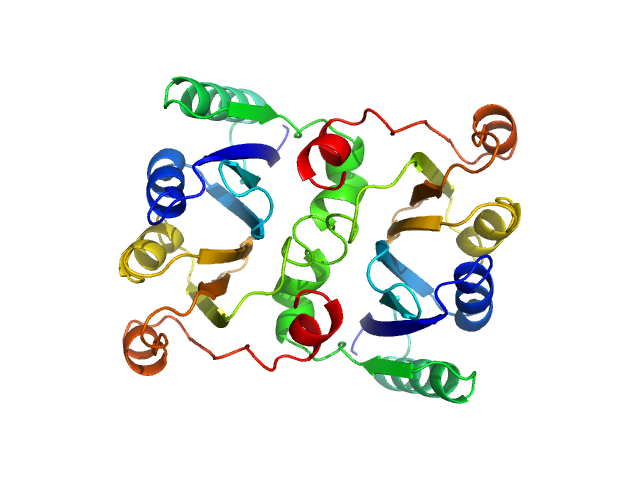
|
| Sample: |
L-threonine dehydratase biosynthetic IlvA Regulatory domain dimer, 46 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM KPO4, 200 mM NaCl, 0.1 mM TCEP,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 14
|
Isoleucine Binding and Regulation of Escherichia coli and Staphylococcus aureus Threonine Dehydratase (IlvA).
Biochemistry (2025)
Yun MK, Subramanian C, Miller K, Jackson P, Radka CD, Rock CO
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
UniProt ID: Q2FF63 (336-422) L-threonine dehydratase biosynthetic IlvA
|
|
|
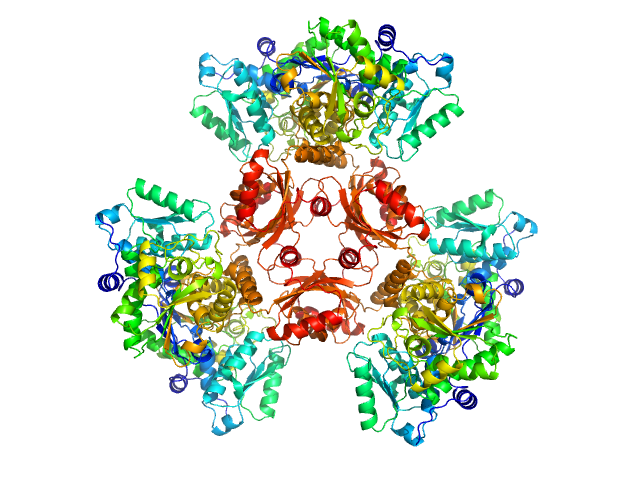
|
| Sample: |
L-threonine dehydratase biosynthetic IlvA hexamer, 288 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
20 mM KPO4, 200 mM NaCl, 0.1 mM TCEP,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 14
|
Isoleucine Binding and Regulation of Escherichia coli and Staphylococcus aureus Threonine Dehydratase (IlvA).
Biochemistry (2025)
Yun MK, Subramanian C, Miller K, Jackson P, Radka CD, Rock CO
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
386 |
nm3 |
|
|
UniProt ID: P55265 (688-817) Third double-stranded RNA-binding domain of human ADAR1 (residues 688-817, i.e. dsRBD3-long)
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (residues 688-817, i.e. dsRBD3-long) dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES, 55 mM KOAc, 10 mM NaCl, 1 mM TCEP, pH: 7.3 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 27
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
UniProt ID: P55265 (708-801) Third double-stranded RNA-binding domain of human ADAR1 (residues 708-801, i.e. dsRBD3-mid)
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (residues 708-801, i.e. dsRBD3-mid) dimer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES, 55 mM KOAc, 10 mM NaCl, 1 mM TCEP, pH: 7.3 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 27
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
UniProt ID: P55265 (716-797) Third double-stranded RNA-binding domain of human ADAR1 (residues 716-797, i.e. dsRBD3-short)
|
|
|
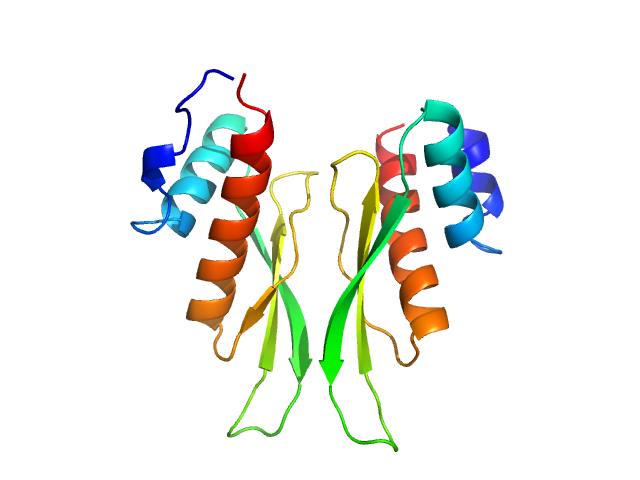
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (residues 716-797, i.e. dsRBD3-short) dimer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES, 55 mM KOAc, 10 mM NaCl, 1 mM TCEP, pH: 7.3 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 27
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
UniProt ID: P55265 (708-801) Third double-stranded RNA-binding domain of human ADAR1 (wild-type, residues 708-801, i.e. ADAR1-dsRBD3)
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (wild-type, residues 708-801, i.e. ADAR1-dsRBD3) dimer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-phosphate, 100 mM NaCl, 2 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 8
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
UniProt ID: P55265 (708-801) Third double-stranded RNA-binding domain of human ADAR1 (interface mutant, residues 708-801, i.e. ADAR1-dsRBD3 interface mutant)
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (interface mutant, residues 708-801, i.e. ADAR1-dsRBD3 interface mutant) monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-phosphate, 100 mM NaCl, 2 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 8
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
UniProt ID: P55265 (708-801) Chimeric construct between the third dsRBD of human ADAR1 and the second dsRBD of Xenopus Xlrbpa (chimeric construct, residues 708-801, i.e. Chimeric ADAR1-ds3/Xlrbpa-ds2)
|
|
|
|
| Sample: |
Chimeric construct between the third dsRBD of human ADAR1 and the second dsRBD of Xenopus Xlrbpa (chimeric construct, residues 708-801, i.e. Chimeric ADAR1-ds3/Xlrbpa-ds2) monomer, 13 kDa Homo sapiens / … protein
|
| Buffer: |
20 mM Na-phosphate, 100 mM NaCl, 2 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 8
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
16 |
nm3 |
|
|