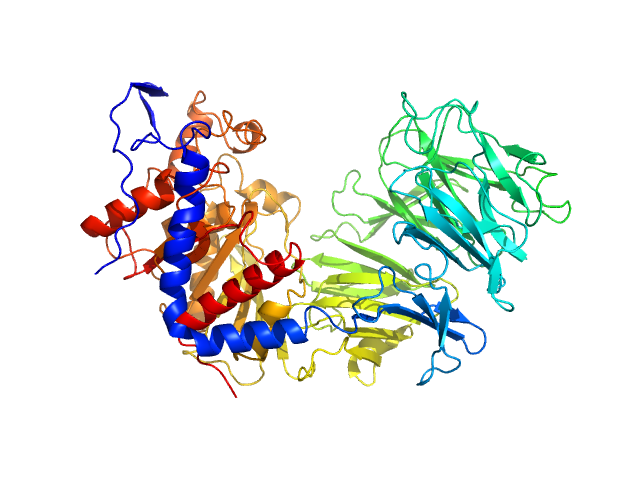
UniProt ID: Q71MD6 (1-697) Prolyl endopeptidase
|
|
|
|
| Sample: |
Prolyl endopeptidase monomer, 78 kDa Trypanosoma cruzi protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Apr 24
|
Cryo-EM led analysis of open and closed conformations of Chagas vaccine candidate TcPOP.
Nat Commun 16(1):7164 (2025)
Batra S, Olmo F, Ragan TJ, Kaplan M, Calvaresi V, Frank AM, Lancey C, Assadipapari M, Ying C, Struwe WB, Hesketh EL, Kelly JM, Barfod L, Campeotto I
|
| RgGuinier |
3.0 |
nm |
| Dmax |
0.0 |
nm |
| VolumePorod |
142 |
nm3 |
|
|
UniProt ID: Q71MD6 (None-None) Prolyl endopeptidase
|
|
|
|
| Sample: |
Prolyl endopeptidase monomer, 78 kDa Trypanosoma cruzi protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Apr 24
|
Cryo-EM led analysis of open and closed conformations of Chagas vaccine candidate TcPOP.
Nat Commun 16(1):7164 (2025)
Batra S, Olmo F, Ragan TJ, Kaplan M, Calvaresi V, Frank AM, Lancey C, Assadipapari M, Ying C, Struwe WB, Hesketh EL, Kelly JM, Barfod L, Campeotto I
|
| RgGuinier |
3.8 |
nm |
| Dmax |
0.0 |
nm |
| VolumePorod |
170 |
nm3 |
|
|
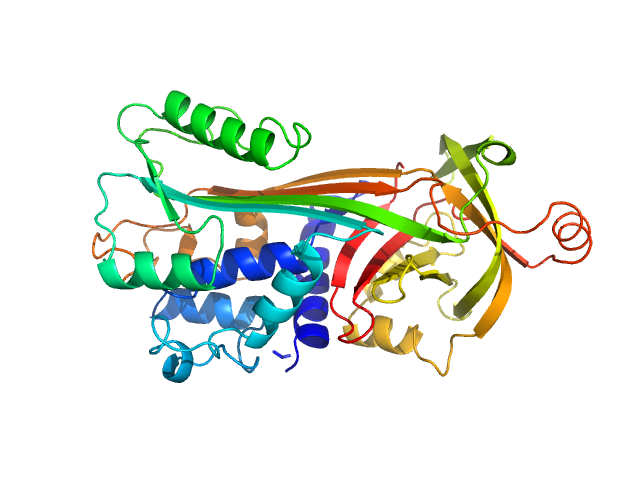
UniProt ID: P01012 (2-386) Ovalbumin
|
|
|
|
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
100 mM Tris-HCl, 100 mM NaCl in 100% v/v D₂O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Apr 22
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.5 |
nm |
|
|
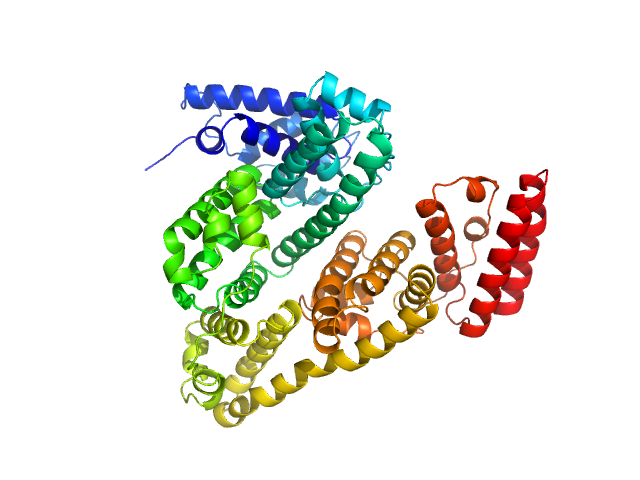
UniProt ID: P02769 (None-None) Albumin (natural variant A214T)
|
|
|
|
| Sample: |
Albumin (natural variant A214T) monomer, 66 kDa Bos taurus protein
|
| Buffer: |
100 mM Tris-HCl, 100 mM NaCl in 100% v/v D₂O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Apr 22
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.7 |
nm |
|
|
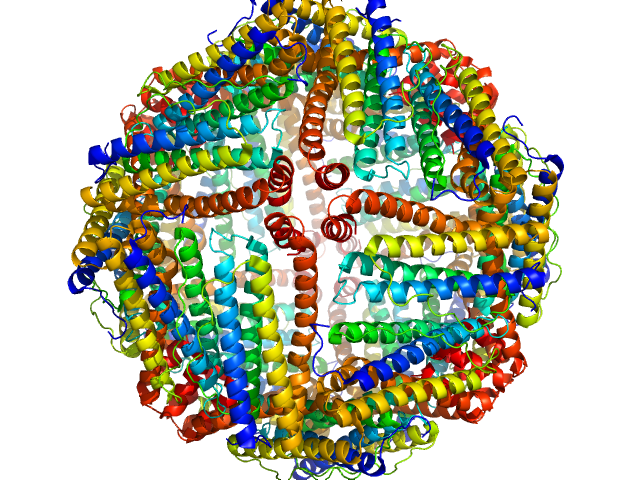
UniProt ID: P02791 (2-175) Ferritin light chain
|
|
|
|
| Sample: |
Ferritin light chain 24-mer, 476 kDa Equus caballus protein
|
| Buffer: |
100 mM Tris-HCl, 100 mM NaCl in 100% v/v D₂O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Apr 22
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
5.4 |
nm |
| Dmax |
23.1 |
nm |
|
|
UniProt ID: Q5N595 (1-102) Circadian clock oscillator protein KaiB
UniProt ID: Q79PF4 (2-519) Circadian clock oscillator protein KaiC
|
|
|
|
| Sample: |
Circadian clock oscillator protein KaiB hexamer, 71 kDa Synechococcus sp. (strain … protein
Circadian clock oscillator protein KaiC hexamer, 356 kDa Synechococcus elongatus (strain … protein
|
| Buffer: |
50 mM sodium phosphate buffer, 150 mM NaCl, 5 mM MgCl₂, 0.5 mM EDTA, 1 mM ATP, 1 mM DTT, 50 mM arginine, 50 mM glutamine, in 100% D₂O, pH: 7.8 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Jul 2
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
4.9 |
nm |
| Dmax |
14.4 |
nm |
|
|
UniProt ID: Q5N595 (1-102) Circadian clock oscillator protein KaiB
UniProt ID: Q79PF4 (2-519) Circadian clock oscillator protein KaiC
|
|
|
|
| Sample: |
Circadian clock oscillator protein KaiB hexamer, 71 kDa Synechococcus sp. (strain … protein
Circadian clock oscillator protein KaiC hexamer, 356 kDa Synechococcus elongatus (strain … protein
|
| Buffer: |
50 mM sodium phosphate buffer, 150 mM NaCl, 5 mM MgCl₂, 0.5 mM EDTA, 1 mM ATP, 1 mM DTT, 50 mM arginine, 50 mM glutamine, in 100% D₂O, pH: 7.8 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Jul 2
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
4.3 |
nm |
| Dmax |
11.7 |
nm |
|
|
UniProt ID: Q79PF4 (2-519) Circadian clock oscillator protein KaiC
|
|
|
|
| Sample: |
Circadian clock oscillator protein KaiC hexamer, 356 kDa Synechococcus elongatus (strain … protein
|
| Buffer: |
50 mM sodium phosphate buffer, 150 mM NaCl, 5 mM MgCl₂, 0.5 mM EDTA, 1 mM ATP, 1 mM DTT, 50 mM arginine, 50 mM glutamine, in 100% D₂O, pH: 7.8 |
| Experiment: |
SANS
data collected at SANS-U, JRR-3 (Japan Atomic Energy Agency) on 2024 Jul 2
|
Size-exclusion chromatography–small-angle neutron scattering system optimized for an instrument with medium neutron flux
Journal of Applied Crystallography 58(2) (2025)
Morishima K, Inoue R, Nakagawa T, Shimizu M, Sakamoto R, Oda T, Mayumi K, Sugiyama M
|
| RgGuinier |
4.4 |
nm |
| Dmax |
11.9 |
nm |
|
|
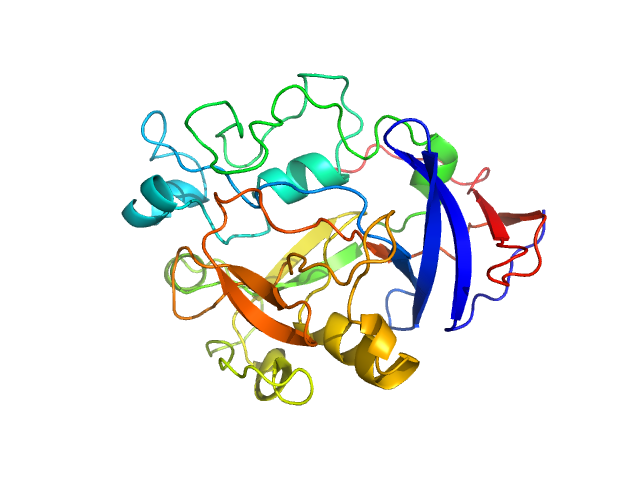
UniProt ID: Q9AES2 (31-313) IgA protease
|
|
|
|
| Sample: |
IgA protease monomer, 32 kDa Thomasclavelia ramosa protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Jun 26
|
Structure of the Thomasclavelia ramosa immunoglobulin A protease reveals a modular and minimizable architecture distinct from other immunoglobulin A proteases.
Proc Natl Acad Sci U S A 122(35):e2503549122 (2025)
Tran N, Frenette A, Holyoak T
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
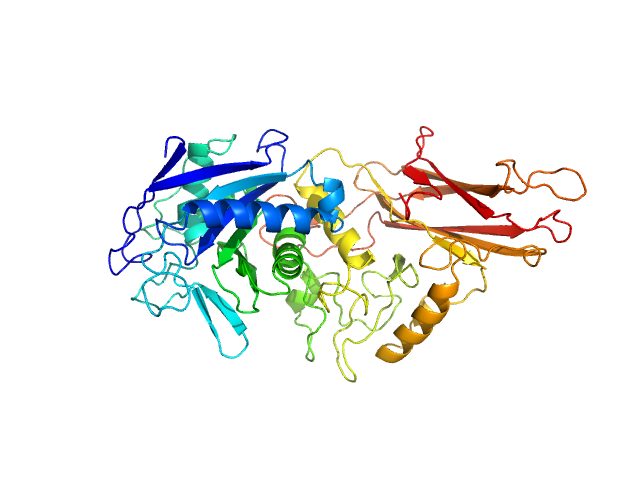
UniProt ID: Q9AES2 (329-807) IgA protease
|
|
|
|
| Sample: |
IgA protease monomer, 55 kDa Thomasclavelia ramosa protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Jun 26
|
Structure of the Thomasclavelia ramosa immunoglobulin A protease reveals a modular and minimizable architecture distinct from other immunoglobulin A proteases.
Proc Natl Acad Sci U S A 122(35):e2503549122 (2025)
Tran N, Frenette A, Holyoak T
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
82 |
nm3 |
|
|