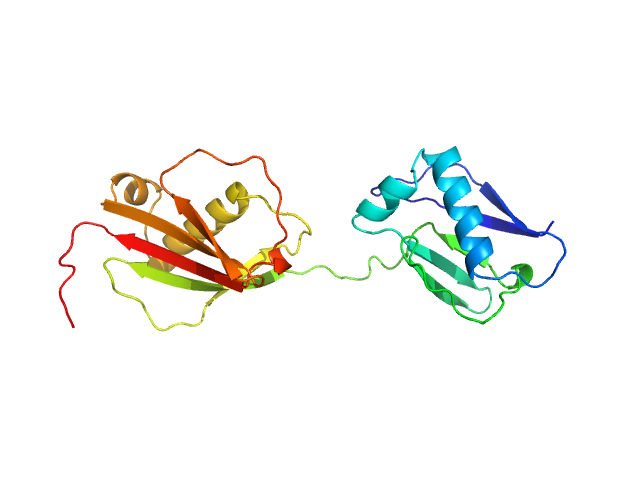
UniProt ID: Q99QS1 (266-476) Protein map
|
|
|
|
| Sample: |
Protein map monomer, 24 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Nov 17
|
S. aureus Eap is a polyvalent inhibitor of neutrophil serine proteases.
J Biol Chem 300(9):107627 (2024)
Mishra N, Gido CD, Herdendorf TJ, Hammel M, Hura GL, Fu ZQ, Geisbrecht BV
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
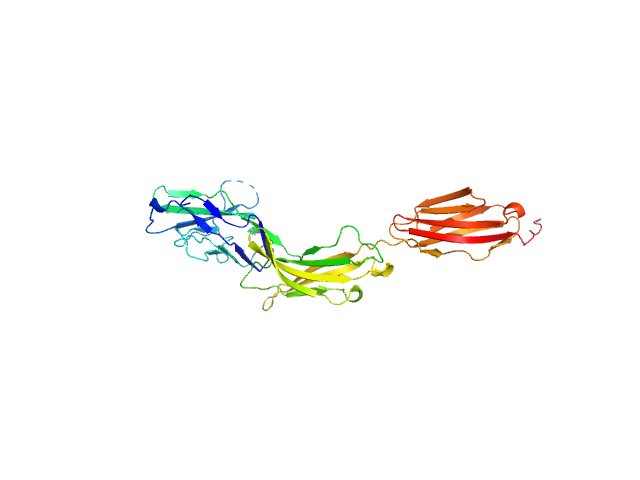
UniProt ID: P20917 (20-325) Myelin-associated glycoprotein Ig domains 1-3
|
|
|
|
| Sample: |
Myelin-associated glycoprotein Ig domains 1-3 monomer, 35 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Sep 11
|
Structural basis of myelin-associated glycoprotein adhesion and signalling.
Nat Commun 7:13584 (2016)
Pronker MF, Lemstra S, Snijder J, Heck AJ, Thies-Weesie DM, Pasterkamp RJ, Janssen BJ
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
UniProt ID: Q46085 (718-1021) Collagenase ColH segement s2as2bs3
|
|
|

|
| Sample: |
Collagenase ColH segement s2as2bs3 monomer, 34 kDa Hathewaya histolytica protein
|
| Buffer: |
10mM HEPES 100mM NaCl 0.2mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ - Induced Structural Change of Multi-Domain Collagen Binding Segments of Collagenases ColG and ColH from Hathewaya histolytica
University of Arkansas Dissertation - (2018)
Christopher E Ruth
|
| RgGuinier |
3.3 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
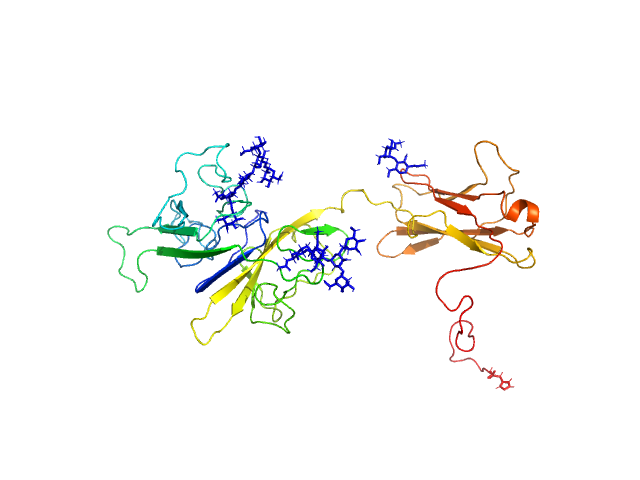
UniProt ID: O95256 (20-364) Interleukin-18 receptor accessory protein ectodomain with Rα linker
|
|
|
|
| Sample: |
Interleukin-18 receptor accessory protein ectodomain with Rα linker monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jul 24
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.4 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
UniProt ID: None (None-None) 80bp_DNA Forward
UniProt ID: None (None-None) 80bp_DNA Reverse
UniProt ID: P0ACF0 (1-90) DNA-binding protein HU-alpha
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, 10 kDa Escherichia coli protein
|
| Buffer: |
10 mM sodium acetate, 100 mM NaCl, pH: 4.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 1
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
|
|
UniProt ID: None (None-None) n-Dodecyl-β-D-Maltopyranoside
UniProt ID: I6YC99 (39-320) Mce-family protein Mce4A
|
|
|

|
| Sample: |
N-Dodecyl-β-D-Maltopyranoside 0, 102 kDa
Mce-family protein Mce4A monomer, 34 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50mM Tris, 500mM NaCl, 10% Glycerol, 5mM DDM, 1mM Beta-ME, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 May 2
|
Structural insights into the substrate-binding proteins Mce1A and Mce4A from Mycobacterium tuberculosis
IUCrJ 8(5) (2021)
Asthana P, Singh D, Pedersen J, Hynönen M, Sulu R, Murthy A, Laitaoja M, Jänis J, Riley L, Venkatesan R
|
| RgGuinier |
4.8 |
nm |
| Dmax |
18.2 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
UniProt ID: O60885 (38-460) Bromodomain-containing protein 4
|
|
|
|
| Sample: |
Bromodomain-containing protein 4 monomer, 48 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 25
|
Multivalent nucleosome scaffolding by bromodomain and extraterminal domain tandem bromodomains.
J Biol Chem :108289 (2025)
Olp MD, Bursch KL, Wynia-Smith SL, Nuñez R, Goetz CJ, Jackson V, Smith BC
|
| RgGuinier |
7.3 |
nm |
| Dmax |
20.2 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
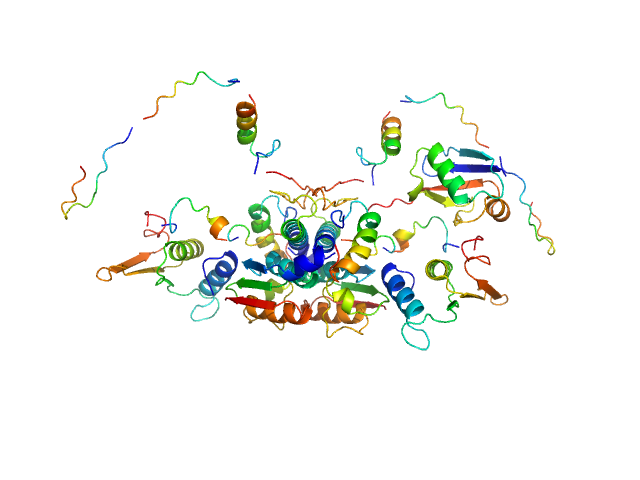
UniProt ID: Q9GZZ9 (57-346) Ubiquitin-like modifier-activating enzyme 5
UniProt ID: P61960 (1-83) Ubiquitin fold modifer 1
|
|
|
|
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
Ubiquitin fold modifer 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 29
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
UniProt ID: P02768 (None-None) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.4 |
nm |
|
|
UniProt ID: P0DN75 (2-132) Iron-sulfur cluster assembly 1 homolog, mitochondrial
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Magnetic field effects on the structure and molecular behavior of pigeon iron–sulfur protein
Protein Science 31(6) (2022)
Arai S, Shimizu R, Adachi M, Hirai M
|
| RgGuinier |
4.7 |
nm |
| Dmax |
15.8 |
nm |
|
|