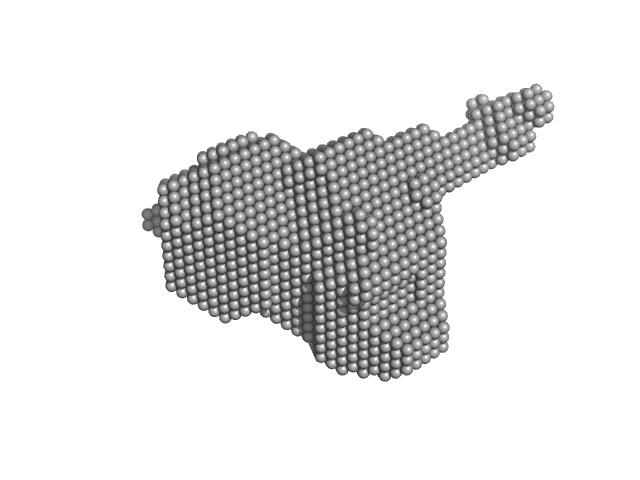
UniProt ID: P74102 (2-317) Orange carotenoid-binding protein
|
|
|
|
| Sample: |
Orange carotenoid-binding protein monomer, 35 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
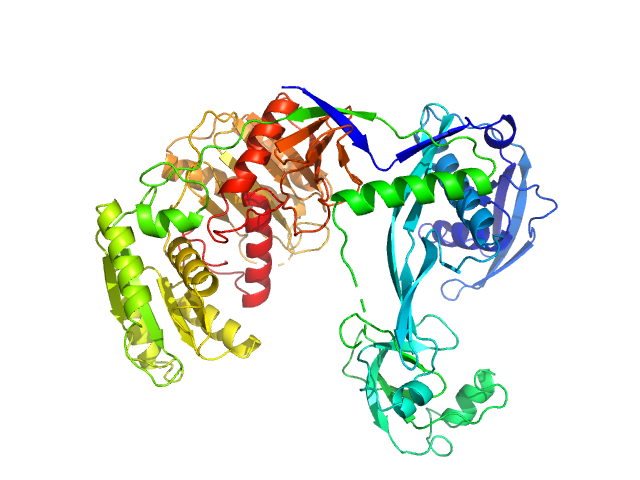
UniProt ID: Q746M7 (1-685) Piwi domain-containing protein
|
|
|
|
| Sample: |
Piwi domain-containing protein monomer, 77 kDa Thermus thermophilus (strain … protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
UniProt ID: P0DTC9 (44-180) Nucleoprotein
UniProt ID: None (None-None) AU extension in the 5'-genomic end of SARS-CoV-2
|
|
|
|
| Sample: |
Nucleoprotein monomer, 15 kDa Severe acute respiratory … protein
AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 7 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Aug 16
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
2.1 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
UniProt ID: P73129 (1-96) Ssr1698 protein (H21A)
|
|
|
|
| Sample: |
Ssr1698 protein (H21A) dimer, 22 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2022 Dec 7
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
UniProt ID: P0DTC9 (1-245) Nucleoprotein
|
|
|
|
| Sample: |
Nucleoprotein dimer, 55 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
SARS-CoV-2 N-protein variants: N1-246 and IDL176-246
Guillem Hernandez
|
| RgGuinier |
4.0 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
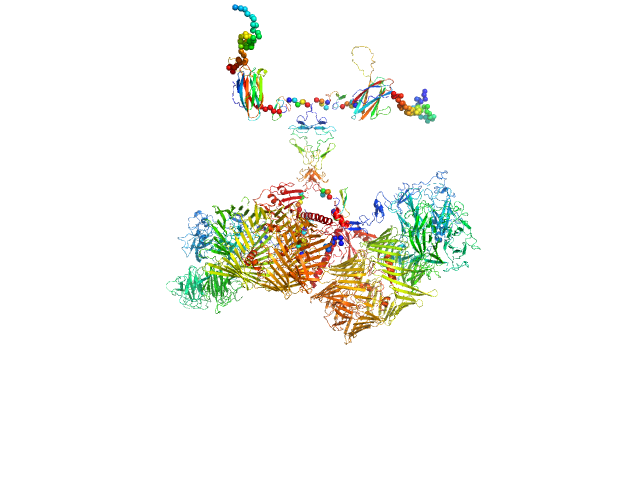
UniProt ID: Q9WTS6-2 (343-2699) Isoform A0B0 of Teneurin-3
|
|
|
|
| Sample: |
Isoform A0B0 of Teneurin-3 dimer, 526 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
7.3 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
1057 |
nm3 |
|
|
UniProt ID: P08311 (22-245) Cathepsin G
UniProt ID: P08246 (31-249) Neutrophil elastase
UniProt ID: Q99QS1 (268-438) Eap34
|
|
|
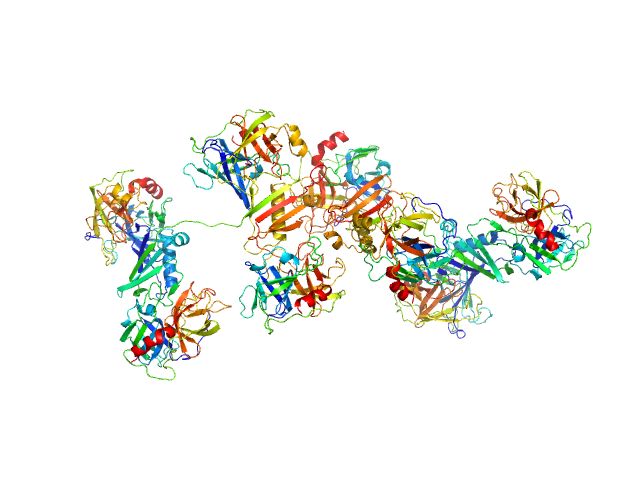
|
| Sample: |
Cathepsin G tetramer, 101 kDa Homo sapiens protein
Neutrophil elastase tetramer, 93 kDa Homo sapiens protein
Eap34 dimer, 48 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
20 mM HEPES, 140 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Jun 3
|
S. aureus Eap is a polyvalent inhibitor of neutrophil serine proteases.
J Biol Chem 300(9):107627 (2024)
Mishra N, Gido CD, Herdendorf TJ, Hammel M, Hura GL, Fu ZQ, Geisbrecht BV
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.4 |
nm |
| VolumePorod |
500 |
nm3 |
|
|
UniProt ID: Q46085 (718-1021) Collagenase ColH segement s2as2bs3
|
|
|
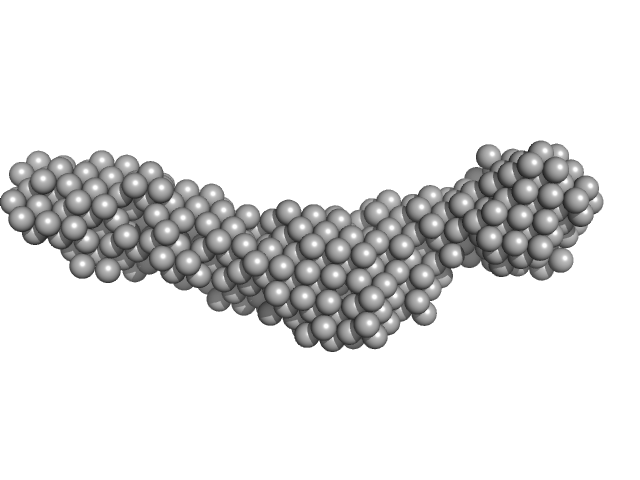
|
| Sample: |
Collagenase ColH segement s2as2bs3 monomer, 34 kDa Hathewaya histolytica protein
|
| Buffer: |
10mM HEPES 100mM NaCl 0.2mM EGTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Oct 12
|
Ca2+ - Induced Structural Change of Multi-Domain Collagen Binding Segments of Collagenases ColG and ColH from Hathewaya histolytica
University of Arkansas Dissertation - (2018)
Christopher E Ruth
|
| RgGuinier |
3.2 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
UniProt ID: None (None-None) 80bp_DNA Forward
UniProt ID: None (None-None) 80bp_DNA Reverse
UniProt ID: P0ACF0 (1-90) DNA-binding protein HU-alpha
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, 10 kDa Escherichia coli protein
|
| Buffer: |
10 mM sodium acetate, 150 mM NaCl, pH: 4.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Jun 1
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
|
|
UniProt ID: Q9GZZ9 (57-346) Ubiquitin-like modifier-activating enzyme 5
|
|
|
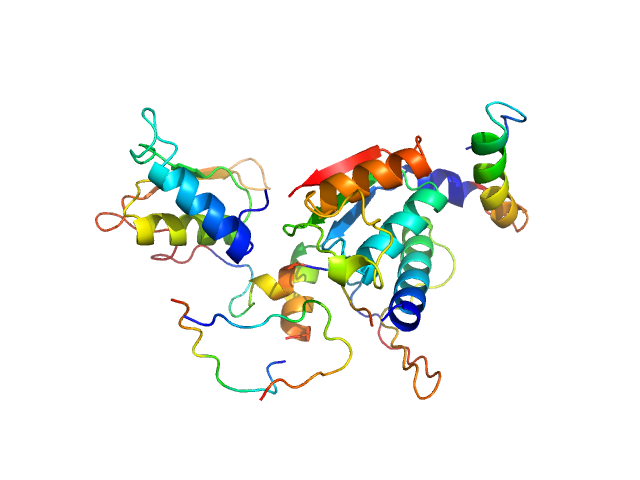
|
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 13
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
74 |
nm3 |
|
|