UniProt ID: P74103 (8-109) Fluorescence recovery protein
|
|
|

|
| Sample: |
Fluorescence recovery protein dimer, 23 kDa Synechocystis sp. PCC … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.1 mM EDTA, 2 mM dithiothreitol, 3 % v/v glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
Functional interaction of low-homology FRPs from different cyanobacteria with Synechocystis OCP.
Biochim Biophys Acta 1859(5):382-393 (2018)
Slonimskiy YB, Maksimov EG, Lukashev EP, Moldenhauer M, Jeffries CM, Svergun DI, Friedrich T, Sluchanko NN
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
UniProt ID: B5W3T4 (1-106) Uncharacterized fluorescence recovery protein
|
|
|

|
| Sample: |
Uncharacterized fluorescence recovery protein dimer, 24 kDa Arthrospira maxima CS-328 protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.1 mM EDTA, 2 mM dithiothreitol, 3 % v/v glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 1
|
Functional interaction of low-homology FRPs from different cyanobacteria with Synechocystis OCP.
Biochim Biophys Acta 1859(5):382-393 (2018)
Slonimskiy YB, Maksimov EG, Lukashev EP, Moldenhauer M, Jeffries CM, Svergun DI, Friedrich T, Sluchanko NN
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
UniProt ID: P15146-3 (None-None) Microtubule-associated protein 2, isoform 3
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
50 mM MOPS, 150 mM NaCl, 0.03% NaN3, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
UniProt ID: P15146-3 (None-None) Microtubule-associated protein 2, isoform 3
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
50 mM MOPS, 150 mM NaCl, 0.03% NaN3, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
UniProt ID: P15146-3 (None-None) Microtubule-associated protein 2, isoform 3
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
50 mM MOPS, 150 mM NaCl, 0.03% NaN3, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
UniProt ID: P15146-3 (None-None) Microtubule-associated protein 2, isoform 3
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
50 mM MOPS, 150 mM NaCl, 0.03% NaN3, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
UniProt ID: P15146-3 (None-None) Microtubule-associated protein 2, isoform 3
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
50 mM MOPS, 150 mM NaCl, 0.03% NaN3, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
UniProt ID: P15146-3 (None-None) Microtubule-associated protein 2, isoform 3
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
50 mM MOPS, 150 mM NaCl, 0.03% NaN3, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
UniProt ID: P15146-3 (None-None) Microtubule-associated protein 2, isoform 3
|
|
|
|
| Sample: |
Microtubule-associated protein 2, isoform 3 monomer, 49 kDa Rattus norvegicus protein
|
| Buffer: |
MOPS map2c buffer, pH: 6.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 21
|
Functionally specific binding regions of microtubule-associated protein 2c exhibit distinct conformations and dynamics.
J Biol Chem 293(34):13297-13309 (2018)
Melková K, Zapletal V, Jansen S, Nomilner E, Zachrdla M, Hritz J, Nováček J, Zweckstetter M, Jensen MR, Blackledge M, Žídek L
|
|
|
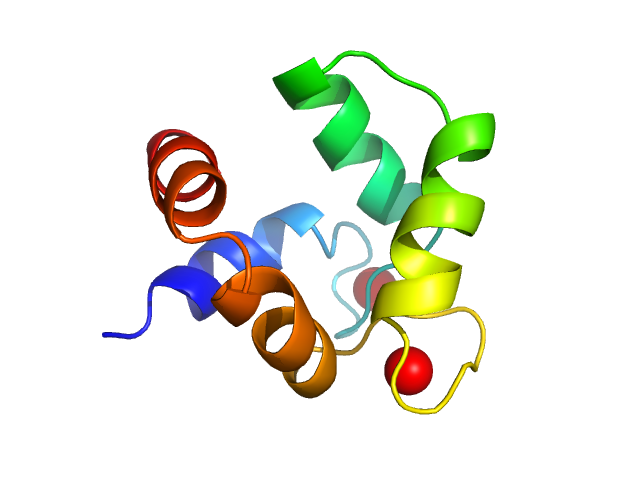
UniProt ID: O82040 (None-None) polcalcin Phl p 7
|
|
|
|
| Sample: |
Polcalcin Phl p 7 monomer, 9 kDa Phleum pratense protein
|
| Buffer: |
0.15M NaCl, 0.025M Hepes, pH 7.4, 100 uM Ca2+, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2011 Dec 8
|
Solution structures of polcalcin Phl p 7 in three ligation states: Apo-, hemi-Mg2+-bound, and fully Ca2+-bound.
Proteins 81(2):300-15 (2013)
Henzl MT, Sirianni AG, Wycoff WG, Tan A, Tanner JJ
|
| RgGuinier |
1.3 |
nm |
| Dmax |
3.6 |
nm |
| VolumePorod |
14 |
nm3 |
|
|