UniProt ID: Q88E39 (None-None) Sensory box protein light-state (R66I)
|
|
|
|
| Sample: |
Sensory box protein light-state (R66I) dimer, 37 kDa Pseudomonas putida protein
|
| Buffer: |
10mM Tris, 10 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Dec 2
|
Small-angle X-ray scattering study of the kinetics of light-dark transition in a LOV protein.
PLoS One 13(7):e0200746 (2018)
Röllen K, Granzin J, Batra-Safferling R, Stadler AM
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
UniProt ID: P02687 (None-None) Myelin basic protein
|
|
|
|
| Sample: |
Myelin basic protein monomer, 18 kDa Bos taurus protein
|
| Buffer: |
20 mM NaH2PO4/ Na2HPO4, 99.9% D2O, pH: 4.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Feb 21
|
Internal nanosecond dynamics in the intrinsically disordered myelin basic protein.
J Am Chem Soc 136(19):6987-94 (2014)
Stadler AM, Stingaciu L, Radulescu A, Holderer O, Monkenbusch M, Biehl R, Richter D
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.1 |
nm |
|
|
UniProt ID: Q88E39 (None-None) Sensory box protein dark-state
|
|
|
|
| Sample: |
Sensory box protein dark-state dimer, 37 kDa Pseudomonas putida protein
|
| Buffer: |
10mM Tris, 10 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Dec 2
|
Small-angle X-ray scattering study of the kinetics of light-dark transition in a LOV protein.
PLoS One 13(7):e0200746 (2018)
Röllen K, Granzin J, Batra-Safferling R, Stadler AM
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
UniProt ID: Q5W264 (4-191) Di-domain acyl carrier protein of PigH from prodigiosin biosynthesis
|
|
|

|
| Sample: |
Di-domain acyl carrier protein of PigH from prodigiosin biosynthesis monomer, 22 kDa Serratia sp. ATCC … protein
|
| Buffer: |
20 mM Tris supplemented with 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2016 Jan 13
|
Solution Structure and Conformational Dynamics of a Doublet Acyl Carrier Protein from Prodigiosin Biosynthesis.
Biochemistry (2021)
Thongkawphueak T, Winter AJ, Williams C, Maple HJ, Soontaranon S, Kaewhan C, Campopiano DJ, Crump MP, Wattana-Amorn P
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
UniProt ID: K5B7F3 (1-219) Methyltransferase domain protein
|
|
|
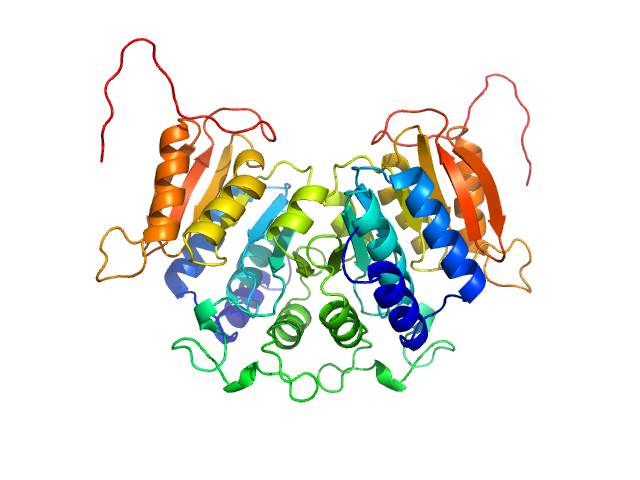
|
| Sample: |
Methyltransferase domain protein dimer, 51 kDa Mycolicibacterium hassiacum protein
|
| Buffer: |
20 mM bis-tris propane, 50 mM NaCl, 2 mM DTT, 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jun 21
|
Biosynthesis of mycobacterial methylmannose polysaccharides requires a unique 1-O-methyltransferase specific for 3-O-methylated mannosides.
Proc Natl Acad Sci U S A 116(3):835-844 (2019)
Ripoll-Rozada J, Costa M, Manso JA, Maranha A, Miranda V, Sequeira A, Ventura MR, Macedo-Ribeiro S, Pereira PJB, Empadinhas N
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
86 |
nm3 |
|
|
UniProt ID: P05937 (None-None) Calbindin
|
|
|
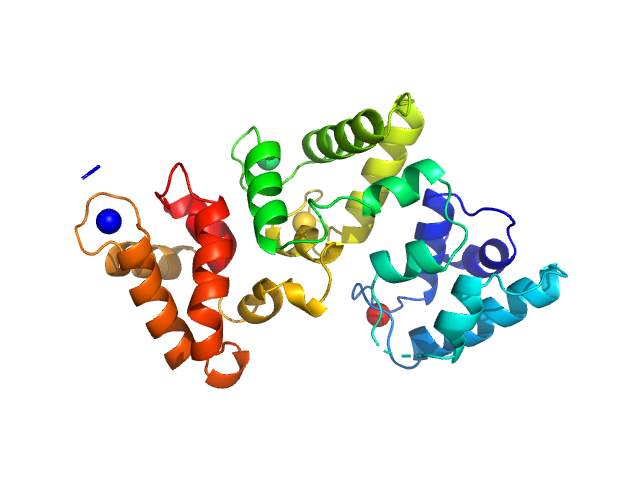
|
| Sample: |
Calbindin monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Feb 28
|
The X-ray structure of human calbindin-D28K: an improved model.
Acta Crystallogr D Struct Biol 74(Pt 10):1008-1014 (2018)
Noble JW, Almalki R, Roe SM, Wagner A, Duman R, Atack JR
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
UniProt ID: P05937 (None-None) Calbindin
|
|
|

|
| Sample: |
Calbindin monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Feb 28
|
The X-ray structure of human calbindin-D28K: an improved model.
Acta Crystallogr D Struct Biol 74(Pt 10):1008-1014 (2018)
Noble JW, Almalki R, Roe SM, Wagner A, Duman R, Atack JR
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
UniProt ID: P0DMV8 (None-None) Heat shock 70 kDa protein 1
|
|
|
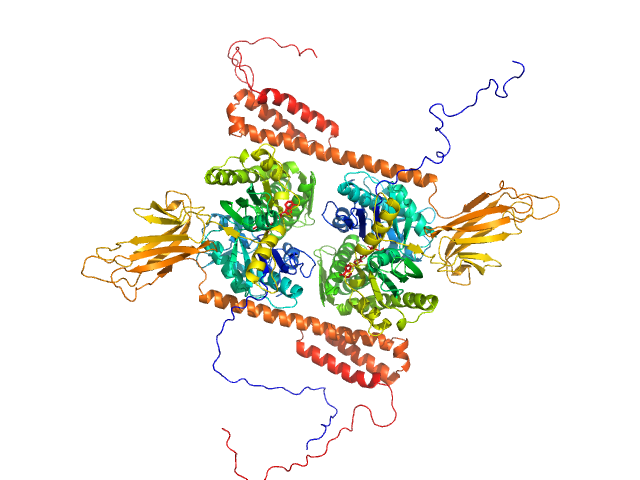
|
| Sample: |
Heat shock 70 kDa protein 1 dimer, 147 kDa Homo sapiens protein
|
| Buffer: |
50mM HEPES, 150mM KCH3COO, 2mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2018 Feb 20
|
Human stress-inducible Hsp70 has a high propensity to form ATP-dependent antiparallel dimers that are differentially regulated by co-chaperone binding.
Mol Cell Proteomics (2018)
Trcka F, Durech M, Vankova P, Chmelik J, Martinkova V, Hausner J, Kadek A, Marcoux J, Klumpler T, Vojtesek B, Muller P, Man P
|
| RgGuinier |
4.0 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
248 |
nm3 |
|
|
UniProt ID: Q6D8U3 (26-924) Ferredoxin Protease
|
|
|
|
| Sample: |
Ferredoxin Protease monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.03 % NaN3, 5.0 % glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Apr 6
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
152 |
nm3 |
|
|
UniProt ID: Q6D8U3 (26-924) Ferredoxin protease E83A mutant
|
|
|
|
| Sample: |
Ferredoxin protease E83A mutant monomer, 101 kDa Pectobacterium atrosepticum SCRI1043 protein
|
| Buffer: |
20 mM Tris HCl, 150 nM NaCl, 0.02 % NaN3, 5% glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Apr 6
|
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
PLoS Biol 16(8):e2006026 (2018)
Grinter R, Hay ID, Song J, Wang J, Teng D, Dhanesakaran V, Wilksch JJ, Davies MR, Littler D, Beckham SA, Henderson IR, Strugnell RA, Dougan G, Lithgow T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
152 |
nm3 |
|
|