UniProt ID: P0AA25 (2-106) Thioredoxin 1
UniProt ID: None (None-None) Alexa Fluor™ 594 C5 Maleimide
UniProt ID: None (None-None) Alexa Fluor™ 488 C5 Hydroxylamine
|
|
|
|
| Sample: |
Thioredoxin 1 monomer, 12 kDa Escherichia coli protein
Alexa Fluor™ 594 C5 Maleimide monomer, 1 kDa
Alexa Fluor™ 488 C5 Hydroxylamine monomer, 1 kDa
|
| Buffer: |
PBS, 10 mM DTT, 6 M urea, 0.3 M KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 15
|
Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements.
Proc Natl Acad Sci U S A 114(31):E6342-E6351 (2017)
Fuertes G, Banterle N, Ruff KM, Chowdhury A, Mercadante D, Koehler C, Kachala M, Estrada Girona G, Milles S, Mishra A, Onck PR, Gräter F, Esteban-Martín S, Pappu RV, Svergun DI, Lemke EA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
UniProt ID: P52948 (2-150) Nuclear pore complex protein Nup98-Nup96
|
|
|
|
| Sample: |
Nuclear pore complex protein Nup98-Nup96 monomer, 15 kDa Homo sapiens protein
|
| Buffer: |
PBS, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 24
|
Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements.
Proc Natl Acad Sci U S A 114(31):E6342-E6351 (2017)
Fuertes G, Banterle N, Ruff KM, Chowdhury A, Mercadante D, Koehler C, Kachala M, Estrada Girona G, Milles S, Mishra A, Onck PR, Gräter F, Esteban-Martín S, Pappu RV, Svergun DI, Lemke EA
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
UniProt ID: P14907 (2-175) Nucleoporin NSP1
|
|
|
|
| Sample: |
Nucleoporin NSP1 monomer, 18 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
PBS, 10 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 24
|
Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements.
Proc Natl Acad Sci U S A 114(31):E6342-E6351 (2017)
Fuertes G, Banterle N, Ruff KM, Chowdhury A, Mercadante D, Koehler C, Kachala M, Estrada Girona G, Milles S, Mishra A, Onck PR, Gräter F, Esteban-Martín S, Pappu RV, Svergun DI, Lemke EA
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
45 |
nm3 |
|
|
UniProt ID: None (None-None) S48 DNA strand 1
UniProt ID: None (None-None) S48 DNA strand 2
UniProt ID: B7JTH0 (1-116) TubR of the pXO1-like plasmid pBc10987 from B. cereus (Bc-TubR)
|
|
|
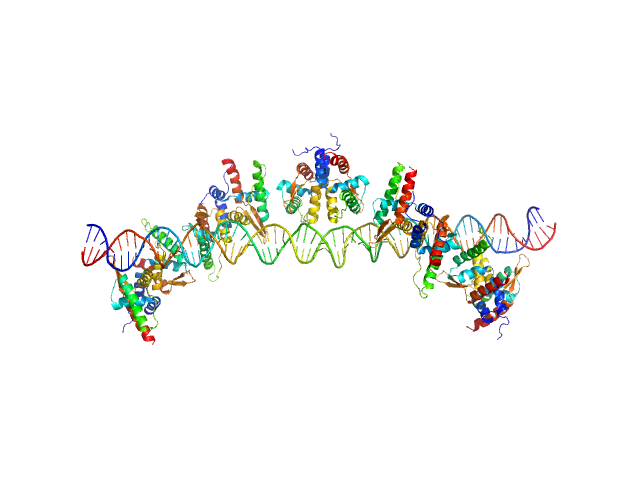
|
| Sample: |
S48 DNA strand 1 monomer, 21 kDa DNA
S48 DNA strand 2 monomer, 21 kDa DNA
TubR of the pXO1-like plasmid pBc10987 from B. cereus (Bc-TubR) decamer, 137 kDa protein
|
| Buffer: |
0.1 M NaCl, 10 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Nov 28
|
Cooperative DNA Binding of the Plasmid Partitioning Protein TubR from the Bacillus cereus pXO1 Plasmid.
J Mol Biol (2018)
Hayashi I, Oda T, Sato M, Fuchigami S
|
| RgGuinier |
6.1 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
305 |
nm3 |
|
|
UniProt ID: P14598 (1-342) Neutophil cytosol factor 1
|
|
|
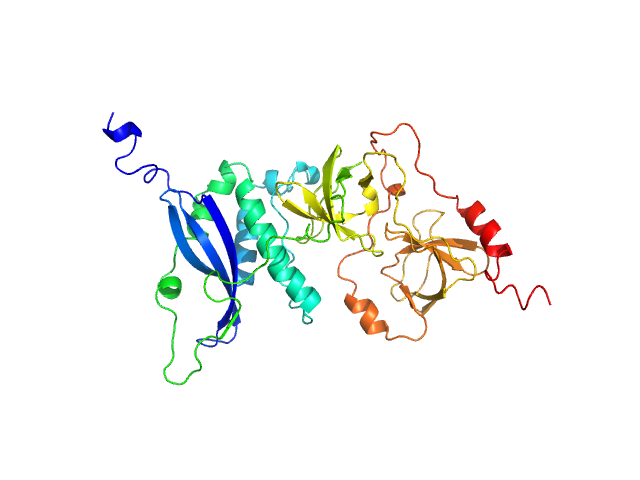
|
| Sample: |
Neutophil cytosol factor 1 monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM EDTA, 2 mM DTT, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, IBBMC on 2009 Oct 16
|
Quantitative live-cell imaging and 3D modeling reveal critical functional features in the cytosolic complex of phagocyte NADPH oxidase.
J Biol Chem (2019)
Ziegler CS, Bouchab L, Tramier M, Durand D, Fieschi F, Dupré-Crochet S, Mérola F, Nüße O, Erard M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
UniProt ID: P14598 (1-390) Neutrophil cytosol factor 1
|
|
|
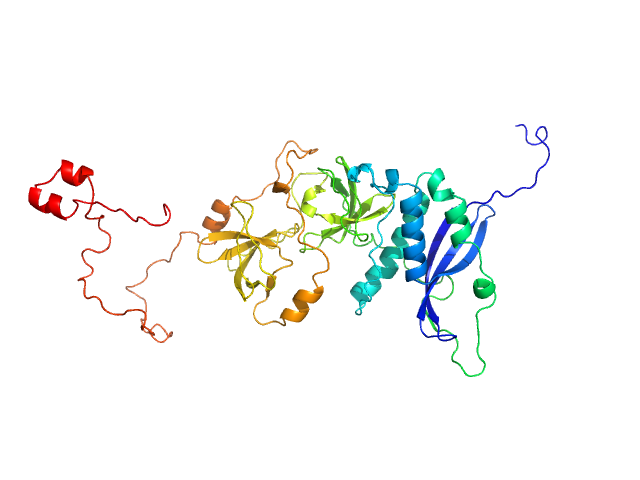
|
| Sample: |
Neutrophil cytosol factor 1 monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM EDTA, 2 mM DTT, 5% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2008 Apr 23
|
Quantitative live-cell imaging and 3D modeling reveal critical functional features in the cytosolic complex of phagocyte NADPH oxidase.
J Biol Chem (2019)
Ziegler CS, Bouchab L, Tramier M, Durand D, Fieschi F, Dupré-Crochet S, Mérola F, Nüße O, Erard M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
UniProt ID: P19878 (1-526) Neutrophil cytosol factor 2
|
|
|
|
| Sample: |
Neutrophil cytosol factor 2 monomer, 61 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 50 mM NaCl, 1 mM EDTA, 2 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at D24, LURE on 2003 Apr 9
|
Quantitative live-cell imaging and 3D modeling reveal critical functional features in the cytosolic complex of phagocyte NADPH oxidase.
J Biol Chem (2019)
Ziegler CS, Bouchab L, Tramier M, Durand D, Fieschi F, Dupré-Crochet S, Mérola F, Nüße O, Erard M
|
| RgGuinier |
4.3 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
UniProt ID: Q6XEC0 (22-265) Beta-lactamase
|
|
|
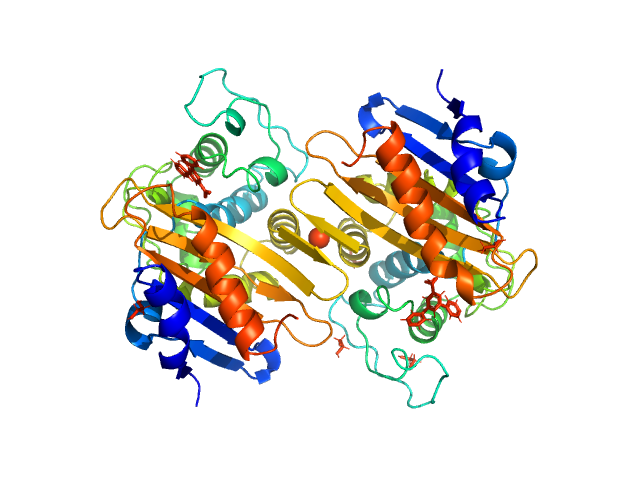
|
| Sample: |
Beta-lactamase dimer, 56 kDa Klebsiella pneumoniae protein
|
| Buffer: |
50 mM HEPES 50 mM K2SO4, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Feb 23
|
The biological assembly of OXA-48 reveals a dimer interface with high charge complementarity and very high affinity.
FEBS J (2018)
Lund BA, Thomassen AM, Nesheim BHB, Carlsen TJO, Isaksson J, Christopeit T, Leiros HS
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
UniProt ID: O77404 (None-None) Tryparedoxin
|
|
|
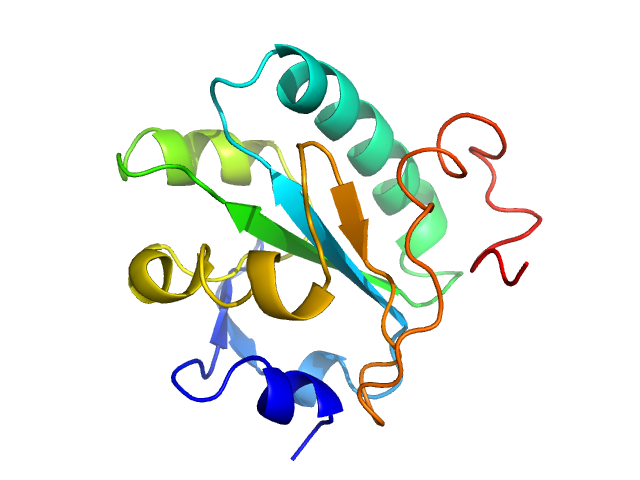
|
| Sample: |
Tryparedoxin monomer, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
UniProt ID: O77404 (None-None) Tryparedoxin
|
|
|

|
| Sample: |
Tryparedoxin monomer, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
27 |
nm3 |
|
|