UniProt ID: (None-None) comcde
UniProt ID: Q8DMW5 (138-250) Response regulator
|
|
|
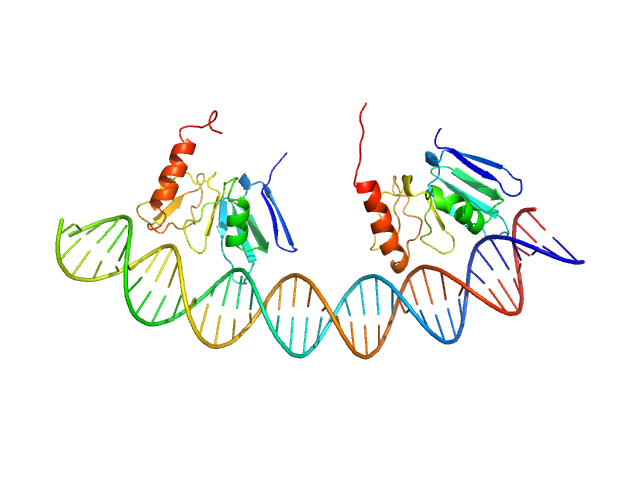
|
| Sample: |
Comcde, 24 kDa Streptococcus pneumoniae DNA
Response regulator dimer, 29 kDa Streptococcus pneumoniae protein
|
| Buffer: |
50 mM MES 500 mM NaCl 5% (v/v) Glycerol 5 mM β-mercaptoethanol, pH: 6.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Jun 2
|
Modeling the ComD/ComE/comcde interaction network using small angle X-ray scattering.
FEBS J 282(8):1538-53 (2015)
Sanchez D, Boudes M, van Tilbeurgh H, Durand D, Quevillon-Cheruel S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
UniProt ID: D9ZNF3 (None-None) Endolysin
|
|
|
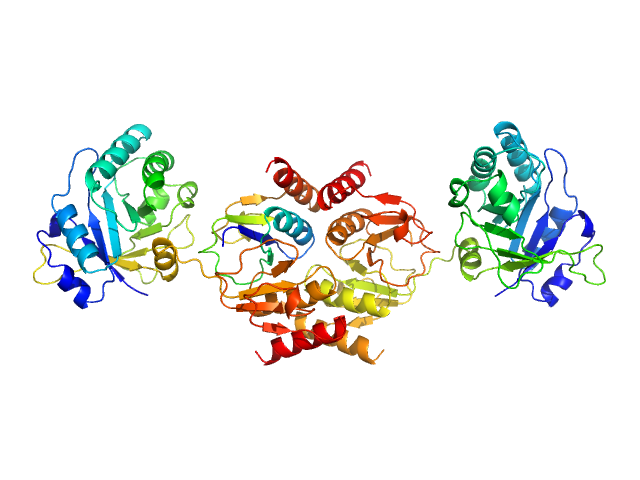
|
| Sample: |
Endolysin , 33 kDa Clostridium phage phiCTP1 protein
|
| Buffer: |
20 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 17
|
Crystal Structure of the CTP1L Endolysin Reveals How Its Activity Is Regulated by a Secondary Translation Product.
J Biol Chem 291(10):4882-93 (2016)
Dunne M, Leicht S, Krichel B, Mertens HD, Thompson A, Krijgsveld J, Svergun DI, Gómez-Torres N, Garde S, Uetrecht C, Narbad A, Mayer MJ, Meijers R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
UniProt ID: I1TJX3 (1-264) Endolysin CS74L
|
|
|
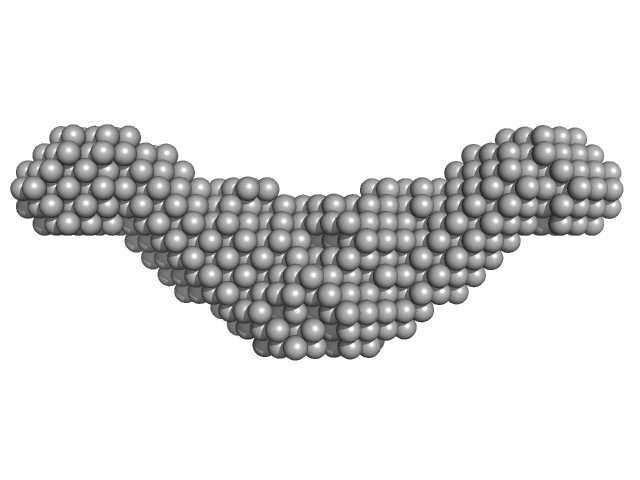
|
| Sample: |
Endolysin CS74L , 31 kDa Clostridium phage phi8074-B1 protein
|
| Buffer: |
20 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 17
|
Crystal Structure of the CTP1L Endolysin Reveals How Its Activity Is Regulated by a Secondary Translation Product.
J Biol Chem 291(10):4882-93 (2016)
Dunne M, Leicht S, Krichel B, Mertens HD, Thompson A, Krijgsveld J, Svergun DI, Gómez-Torres N, Garde S, Uetrecht C, Narbad A, Mayer MJ, Meijers R
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
UniProt ID: O95571 (None-None) Persulfide dioxygenase ETHE1, mitochondrial
|
|
|
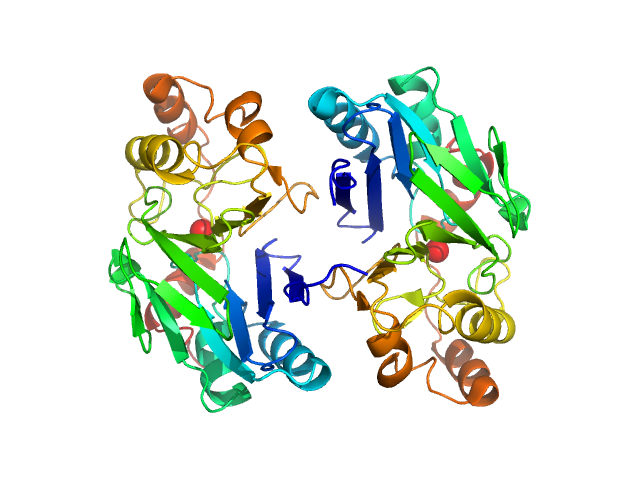
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 21
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
UniProt ID: O95571 (None-None) Persulfide dioxygenase ETHE1, mitochondrial
|
|
|
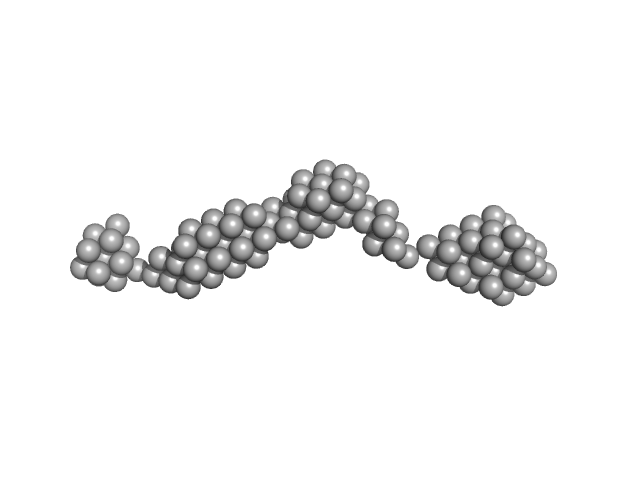
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 23
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
9.0 |
nm |
| Dmax |
32.3 |
nm |
| VolumePorod |
400 |
nm3 |
|
|
UniProt ID: O95571 (None-None) Persulfide dioxygenase ETHE1, mitochondrial
|
|
|
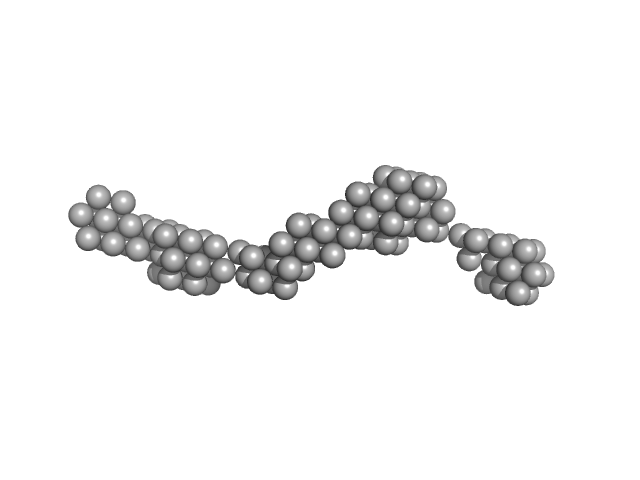
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 23
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
8.3 |
nm |
| Dmax |
38.0 |
nm |
| VolumePorod |
602 |
nm3 |
|
|
UniProt ID: O95571 (None-None) Persulfide dioxygenase ETHE1, mitochondrial
|
|
|
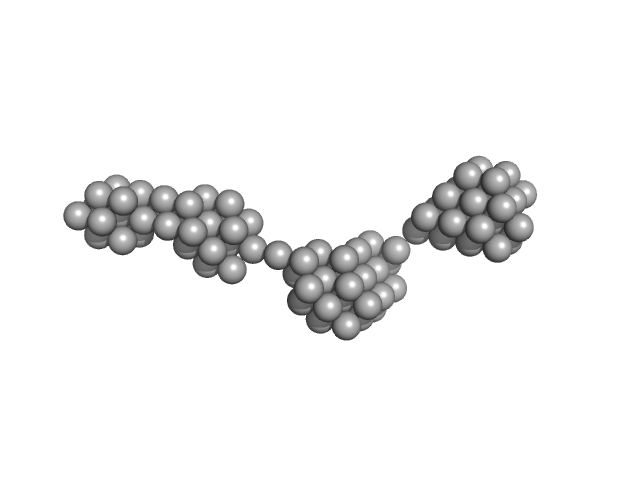
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 23
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
8.8 |
nm |
| Dmax |
42.5 |
nm |
| VolumePorod |
647 |
nm3 |
|
|
UniProt ID: O95571 (None-None) Persulfide dioxygenase ETHE1, mitochondrial
|
|
|

|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 23
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
11.2 |
nm |
| Dmax |
56.0 |
nm |
| VolumePorod |
1445 |
nm3 |
|
|
UniProt ID: O95571 (None-None) Persulfide dioxygenase ETHE1, mitochondrial
|
|
|
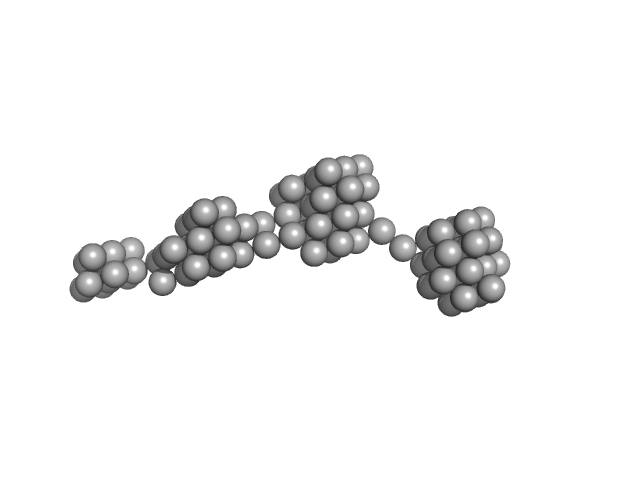
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 23
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
12.4 |
nm |
| Dmax |
59.0 |
nm |
| VolumePorod |
1820 |
nm3 |
|
|
UniProt ID: O95571 (None-None) Persulfide dioxygenase ETHE1, mitochondrial
|
|
|
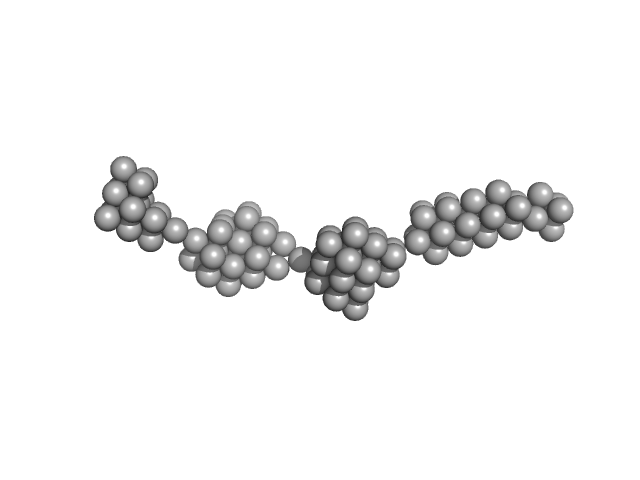
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 23
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
14.2 |
nm |
| Dmax |
63.5 |
nm |
| VolumePorod |
2317 |
nm3 |
|
|