|
|
|
|
|
| Sample: |
Obscurin monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 0.35 mM NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 May 18
|
The N-terminus of obscurin is flexible in solution.
Proteins (2022)
Mauriello GE, Moncure GE, Nowzari RA, Miller CJ, Wright NT
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
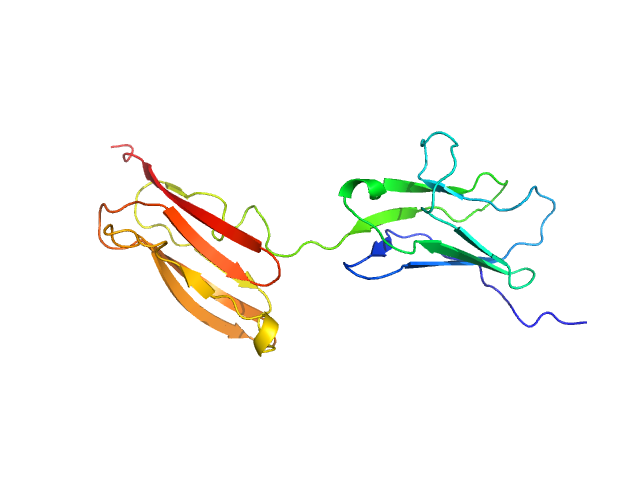
| Sample: |
Obscurin monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 0.35 mM NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Nov 18
|
The N-terminus of obscurin is flexible in solution.
Proteins (2022)
Mauriello GE, Moncure GE, Nowzari RA, Miller CJ, Wright NT
|
| RgGuinier |
3.7 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
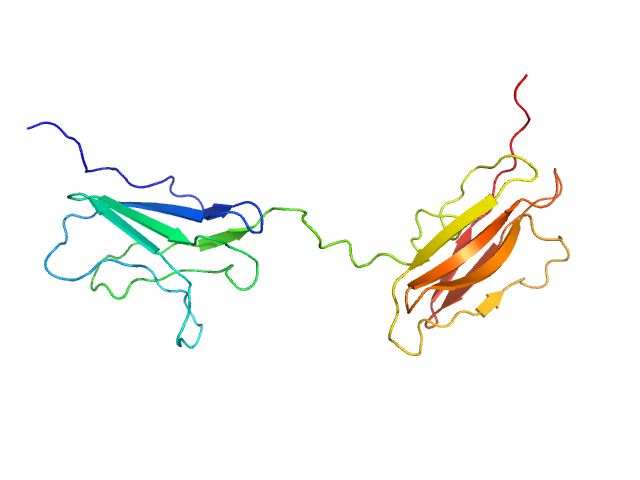
| Sample: |
Obscurin Ig domains 12/13 short monomer, 21 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 0.35 mM NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 20
|
The N-terminus of obscurin is flexible in solution.
Proteins (2022)
Mauriello GE, Moncure GE, Nowzari RA, Miller CJ, Wright NT
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
|
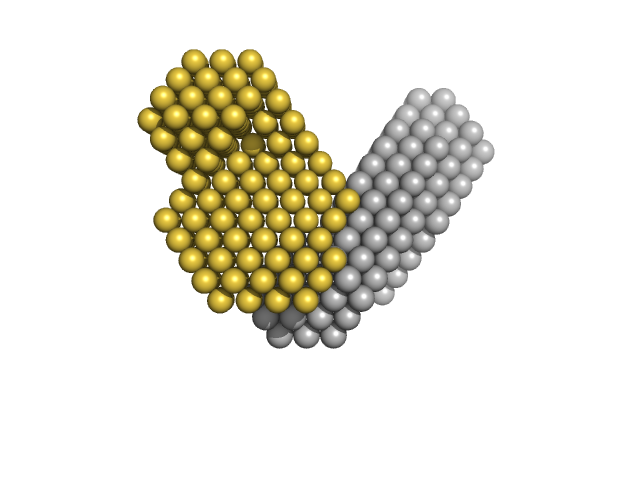
| Sample: |
Obscurin Ig domains 12/13 long monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 0.35 mM NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 20
|
The N-terminus of obscurin is flexible in solution.
Proteins (2022)
Mauriello GE, Moncure GE, Nowzari RA, Miller CJ, Wright NT
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ubiquitin-conjugating enzyme E2 D1 (S22R, C85K, D87S) monomer, 17 kDa Homo sapiens protein
Ubiquitin monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 8 |
| Experiment: |
SANS
data collected at Quokka - Small Angle Neutron Scattering, Australian Centre for Neutron Scattering (ANSTO) on 2019 May 24
|
Production and characterisation of modularly deuterated UBE2D1–Ub conjugate by small angle neutron and X-ray scattering
European Biophysics Journal (2022)
Pietras Z, Duff A, Morad V, Wood K, Jeffries C, Sunnerhagen M
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.4 |
nm |
|
|
|
|
|
|
|
| Sample: |
Phosphoprotein tetramer, 90 kDa Borna disease virus … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM β- mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Sep 1
|
Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses 14(11) (2022)
Tarbouriech N, Chenavier F, Kawasaki J, Bachiri K, Bourhis JM, Legrand P, Freslon LL, Laurent EMN, Suberbielle E, Ruigrok RWH, Tomonaga K, Gonzalez-Dunia D, Horie M, Coyaud E, Crépin T
|
| RgGuinier |
6.1 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
|
|
|
|

|
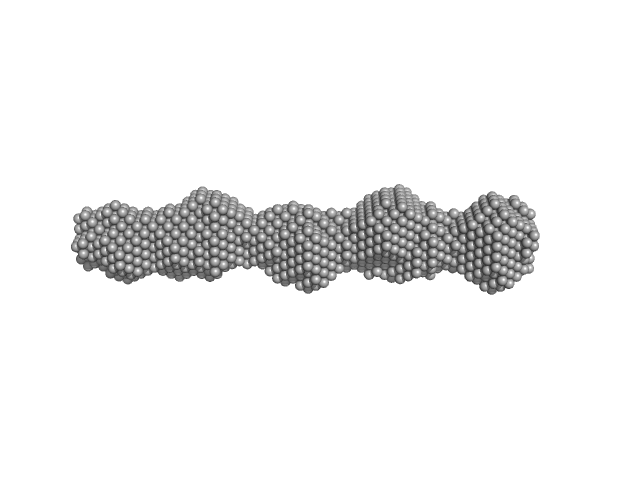
| Sample: |
Phosphoprotein tetramer, 47 kDa Borna disease virus … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM β- mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Sep 1
|
Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses 14(11) (2022)
Tarbouriech N, Chenavier F, Kawasaki J, Bachiri K, Bourhis JM, Legrand P, Freslon LL, Laurent EMN, Suberbielle E, Ruigrok RWH, Tomonaga K, Gonzalez-Dunia D, Horie M, Coyaud E, Crépin T
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphoprotein oligomerisation domain tetramer, 50 kDa protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM β- mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Sep 1
|
Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses 14(11) (2022)
Tarbouriech N, Chenavier F, Kawasaki J, Bachiri K, Bourhis JM, Legrand P, Freslon LL, Laurent EMN, Suberbielle E, Ruigrok RWH, Tomonaga K, Gonzalez-Dunia D, Horie M, Coyaud E, Crépin T
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Phosphoprotein tetramer, 52 kDa Gaboon viper virus … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM β- mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Sep 1
|
Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses 14(11) (2022)
Tarbouriech N, Chenavier F, Kawasaki J, Bachiri K, Bourhis JM, Legrand P, Freslon LL, Laurent EMN, Suberbielle E, Ruigrok RWH, Tomonaga K, Gonzalez-Dunia D, Horie M, Coyaud E, Crépin T
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Skp-like protein trimer, 55 kDa Pseudomonas aeruginosa (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 10 % glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2021 Feb 26
|
The periplasmic chaperone Skp prevents misfolding of the secretory lipase A from Pseudomonas aeruginosa
Frontiers in Molecular Biosciences 9 (2022)
Papadopoulos A, Busch M, Reiners J, Hachani E, Baeumers M, Berger J, Schmitt L, Jaeger K, Kovacic F, Smits S, Kedrov A
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
172 |
nm3 |
|
|