|
|
|
|
|
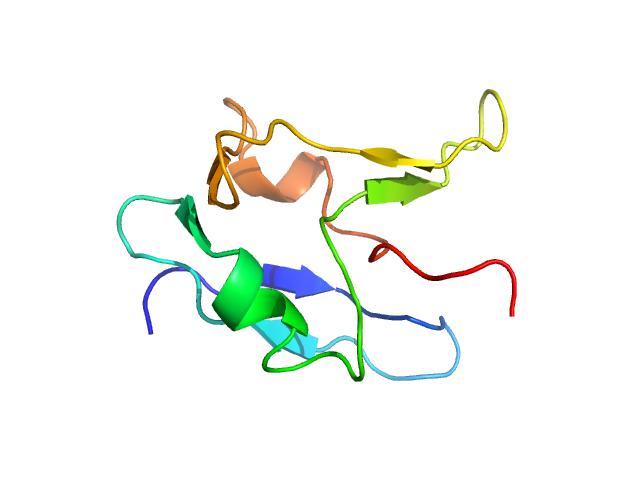
| Sample: |
Cadherin EGF LAG seven-pass G-type receptor 1 dimer, 491 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris, 150 mM NaCl, 1 mM CaCl2, pH: 8.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Dec 8
|
Structural basis for regulation of CELSR1 by a compact module in its extracellular region.
Nat Commun 16(1):3972 (2025)
Bandekar SJ, Garbett K, Kordon SP, Dintzner EE, Li J, Shearer T, Sando RC, Araç D
|
| RgGuinier |
16.2 |
nm |
| Dmax |
67.5 |
nm |
| VolumePorod |
722 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
TAR DNA-binding protein 43 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 3% glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at BL38B1, SPring-8 on 2024 May 29
|
Semi-synthesis of TDP-43 reveals the effects of phosphorylation in N-terminal domain on self-association
Communications Chemistry 8(1) (2025)
Sasaki D, Tenda M, Sohma Y
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
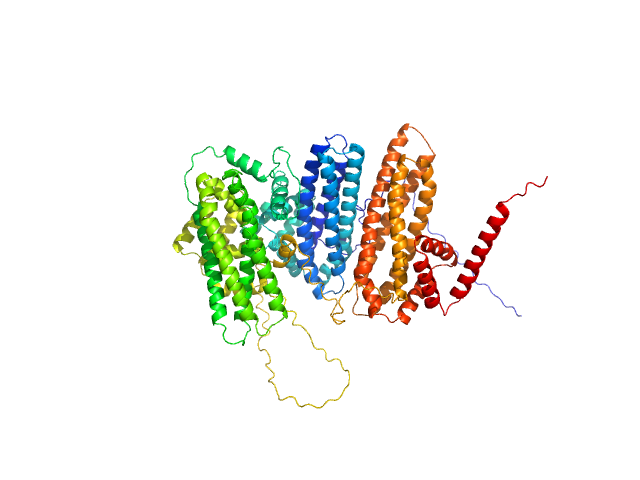
| Sample: |
Zinc finger protein BRUTUS-like At1g18910 monomer, 98 kDa Arabidopsis thaliana protein
|
| Buffer: |
10 mM MES, 15 mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Feb 8
|
Iron-sensing and redox properties of the hemerythrin-like domains of Arabidopsis BRUTUS and BRUTUS-LIKE2 proteins.
Nat Commun 16(1):3865 (2025)
Pullin J, Rodríguez-Celma J, Franceschetti M, Mundy JEA, Svistunenko DA, Bradley JM, Le Brun NE, Balk J
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
|
|
|
|
|
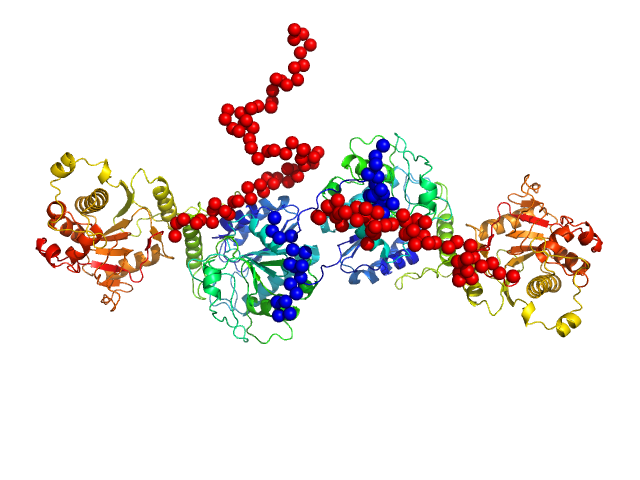
| Sample: |
Procollagen galactosyltransferase 1 dimer, 137 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 0.1 M NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Jun 5
|
Molecular structure and enzymatic mechanism of the human collagen hydroxylysine galactosyltransferase GLT25D1/COLGALT1
Nature Communications 16(1) (2025)
De Marco M, Rai S, Scietti L, Mattoteia D, Liberi S, Moroni E, Pinnola A, Vetrano A, Iacobucci C, Santambrogio C, Colombo G, Forneris F
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
184 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform Tau-F of Microtubule-associated protein tau (C291A, K311C, K317C, C322A) monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
100 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 17
|
Conformational signatures induced by ubiquitin modification in the amyloid-forming tau repeat domain.
Proc Natl Acad Sci U S A 122(15):e2425831122 (2025)
Viola G, Trivellato D, Laitaoja M, Jänis J, Felli IC, D'Onofrio M, Mollica L, Giachin G, Assfalg M
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform Tau-F of Microtubule-associated protein tau (C291A, K311C, K317C, C322A) monomer, 14 kDa Homo sapiens protein
Polyubiquitin-B monomer, 9 kDa Fukomys damarensis protein
|
| Buffer: |
100 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 17
|
Conformational signatures induced by ubiquitin modification in the amyloid-forming tau repeat domain.
Proc Natl Acad Sci U S A 122(15):e2425831122 (2025)
Viola G, Trivellato D, Laitaoja M, Jänis J, Felli IC, D'Onofrio M, Mollica L, Giachin G, Assfalg M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform Tau-F of Microtubule-associated protein tau (C291A, K311C, K317C, C322A) monomer, 14 kDa Homo sapiens protein
Polyubiquitin-B monomer, 9 kDa Fukomys damarensis protein
|
| Buffer: |
100 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 17
|
Conformational signatures induced by ubiquitin modification in the amyloid-forming tau repeat domain.
Proc Natl Acad Sci U S A 122(15):e2425831122 (2025)
Viola G, Trivellato D, Laitaoja M, Jänis J, Felli IC, D'Onofrio M, Mollica L, Giachin G, Assfalg M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform Tau-F of Microtubule-associated protein tau (C291A, K311C, K317C, C322A) monomer, 14 kDa Homo sapiens protein
Polyubiquitin-B monomer, 9 kDa Fukomys damarensis protein
Polyubiquitin-B monomer, 9 kDa Fukomys damarensis protein
|
| Buffer: |
100 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 17
|
Conformational signatures induced by ubiquitin modification in the amyloid-forming tau repeat domain.
Proc Natl Acad Sci U S A 122(15):e2425831122 (2025)
Viola G, Trivellato D, Laitaoja M, Jänis J, Felli IC, D'Onofrio M, Mollica L, Giachin G, Assfalg M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isthmin-1 AMOP-domain monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Pennsylvania State University on 2023 Jun 21
|
Crystal structure of Isthmin-1 and reassessment of its functional role in pre-adipocyte signaling.
Nat Commun 16(1):3580 (2025)
Li T, Stayrook SE, Li W, Wang Y, Li H, Zhang J, Liu Y, Klein DE
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
45264 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
L-AMOP-IFEE monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Pennsylvania State University on 2023 Jun 21
|
Crystal structure of Isthmin-1 and reassessment of its functional role in pre-adipocyte signaling.
Nat Commun 16(1):3580 (2025)
Li T, Stayrook SE, Li W, Wang Y, Li H, Zhang J, Liu Y, Klein DE
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
46874 |
nm3 |
|
|