|
|
|
|

|
| Sample: |
HBV G-quad G1738A monomer, 7 kDa Hepatitis B virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 14
|
HBV G-quad
Trushar Patel
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.3 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
HBV G4 G1748A monomer, 7 kDa Hepatitis B virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 14
|
HBV G-quad
Trushar Patel
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
HBV G4 G1748A + G1748A monomer, 7 kDa Hepatitis B virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, 1 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 17
|
HBV G-quad
Trushar Patel
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ribosome maturation factor RimP monomer, 18 kDa Staphylococcus aureus (strain … protein
30S ribosomal protein S12 monomer, 15 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
50 mM sodium phosphate, 200 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at Xeuss 3.0 SAXS/WAXS System, JINR on 2023 Feb 16
|
Structural aspects of RimP binding on small ribosomal subunit from Staphylococcus aureus.
Structure (2023)
Garaeva N, Fatkhullin B, Murzakhanov F, Gafurov M, Golubev A, Bikmullin A, Glazyrin M, Kieffer B, Jenner L, Klochkov V, Aganov A, Rogachev A, Ivankov O, Validov S, Yusupov M, Usachev K
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
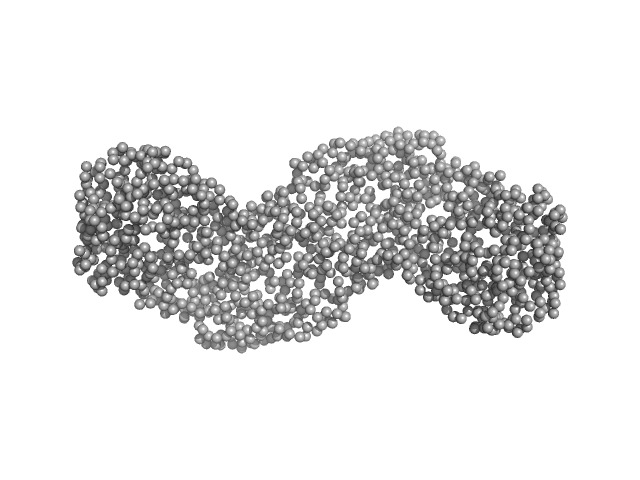
| Sample: |
Gbp6 protein (Gbp7 - Gbp6 protein Mus musculus) dimer, 148 kDa Mus musculus protein
|
| Buffer: |
50 mM Tris, 5 mM MgCl, 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2019 Oct 2
|
Integrative modeling of guanylate binding protein dimers
Protein Science (2023)
Schumann W, Loschwitz J, Reiners J, Degrandi D, Legewie L, Stühler K, Pfeffer K, Poschmann G, Smits S, Strodel B
|
| RgGuinier |
5.4 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
176 |
nm3 |
|
|
|
|
|
|
|
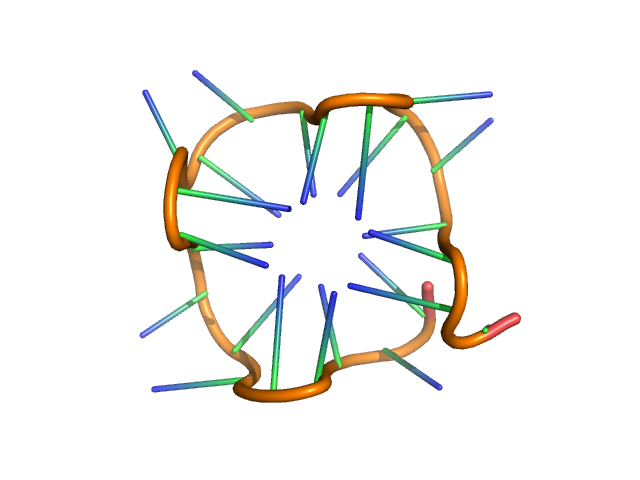
| Sample: |
PolyGU RNA - (GU)12 monomer, 8 kDa synthetic RNA RNA
|
| Buffer: |
20 mM HEPES, 150 mM KCl, pH: 7 |
| Experiment: |
SAXS
data collected at 12-ID-B, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Jun 10
|
Solution structure of poly(UG) RNA
Journal of Molecular Biology :168340 (2023)
Escobar C, Petersen R, Tonelli M, Fan L, Henzler-Wildman K, Butcher S
|
| RgGuinier |
1.4 |
nm |
| Dmax |
5.1 |
nm |
| VolumePorod |
11 |
nm3 |
|
|
|
|
|
|
|
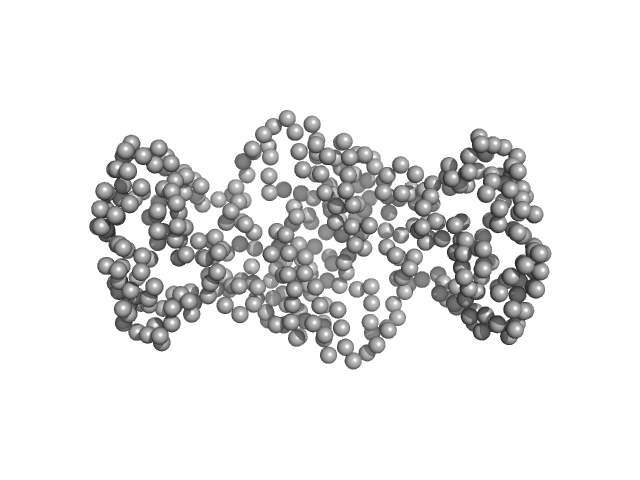
| Sample: |
20 kDa accessory protein dimer, 42 kDa Bacillus thuringiensis serovar … protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2023 Aug 15
|
20-kDa accessory protein (P20) from Bacillus thuringiensis subsp. israelensis ISPC-12: Purification, characterization, solution scattering and structural analysis
International Journal of Biological Macromolecules :127985 (2023)
Kinkar O, Singh R, Prashar A, Kumar A, Hire R, Makde R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Actin, alpha skeletal muscle monomer, 42 kDa Oryctolagus cuniculus protein
|
| Buffer: |
5 mM Tris/Tris-HCl, 0.1 mM CaCl2, 1 mM NaN3, 1.0 mM ATP, 50 mM KCl, 2 mM MgCl2, pH: 8.1 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2021 Jul 1
|
Small-angle X-ray scattering structural insights into alternative pathway of actin oligomerization associated with inactivated state
Biochemical and Biophysical Research Communications :149340 (2023)
Ryzhykau Y, Povarova O, Dronova E, Kuklina D, Antifeeva I, Ilyinsky N, Okhrimenko I, Semenov Y, Kuklin A, Ivanovich V, Fonin A, Uversky V, Turoverov K, Kuznetsova I
|
| RgGuinier |
15.7 |
nm |
| Dmax |
60.0 |
nm |
| VolumePorod |
4710 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDTZ4_fit1_model1.png)
|
| Sample: |
Actin, alpha skeletal muscle monomer, 42 kDa Oryctolagus cuniculus protein
|
| Buffer: |
5 mM Tris/Tris-HCl, 2.0 mM EDTA, pH: 8.1 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2021 Nov 16
|
Small-angle X-ray scattering structural insights into alternative pathway of actin oligomerization associated with inactivated state
Biochemical and Biophysical Research Communications :149340 (2023)
Ryzhykau Y, Povarova O, Dronova E, Kuklina D, Antifeeva I, Ilyinsky N, Okhrimenko I, Semenov Y, Kuklin A, Ivanovich V, Fonin A, Uversky V, Turoverov K, Kuznetsova I
|
| RgGuinier |
4.9 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
549 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDT25_fit1_model1.png)
|
| Sample: |
Actin, alpha skeletal muscle monomer, 42 kDa Oryctolagus cuniculus protein
|
| Buffer: |
5 mM Tris, 0.1 mM CaCl2, 1 mM NaN3, 0.2 mM ATP, pH: 8.1 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2021 Jul 5
|
Small-angle X-ray scattering structural insights into alternative pathway of actin oligomerization associated with inactivated state
Biochemical and Biophysical Research Communications :149340 (2023)
Ryzhykau Y, Povarova O, Dronova E, Kuklina D, Antifeeva I, Ilyinsky N, Okhrimenko I, Semenov Y, Kuklin A, Ivanovich V, Fonin A, Uversky V, Turoverov K, Kuznetsova I
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
84 |
nm3 |
|
|