|
|
|
|

|
| Sample: |
Polyribonucleotide nucleotidyltransferase trimer, 246 kDa Campylobacter jejuni subsp. … protein
|
| Buffer: |
20 mM Tris-HCl, 10 mM NAH2PO4, 60 mM KCl, 1 mM MgCl2, 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Mar 17
|
Structure and function of Campylobacter jejuni polynucleotide phosphorylase (PNPase): Insights into the role of this RNase in pathogenicity.
Biochimie (2023)
Bárria C, Athayde D, Hernandez G, Fonseca L, Casinhas J, Cordeiro TN, Archer M, Arraiano CM, Brito JA, Matos RG
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
310 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Polyribonucleotide nucleotidyltransferase trimer, 237 kDa Campylobacter jejuni subsp. … protein
|
| Buffer: |
20 mM Tris.HCl, 10 mM NAH2PO4, 60 mM KCl, 1 mM MgCl2, 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Mar 17
|
Structure and function of Campylobacter jejuni polynucleotide phosphorylase (PNPase): Insights into the role of this RNase in pathogenicity.
Biochimie (2023)
Bárria C, Athayde D, Hernandez G, Fonseca L, Casinhas J, Cordeiro TN, Archer M, Arraiano CM, Brito JA, Matos RG
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
315 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Polyribonucleotide nucleotidyltransferase trimer, 237 kDa Campylobacter jejuni subsp. … protein
|
| Buffer: |
20 mM Tris.HCl, 10 mM NAH2PO4, 60 mM KCl, 1mM MgCl2, 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Mar 17
|
Structure and function of Campylobacter jejuni polynucleotide phosphorylase (PNPase): Insights into the role of this RNase in pathogenicity.
Biochimie (2023)
Bárria C, Athayde D, Hernandez G, Fonseca L, Casinhas J, Cordeiro TN, Archer M, Arraiano CM, Brito JA, Matos RG
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
312 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-sulfur cluster co-chaperone protein HscB monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 50 mM NaCl, 5 mM KCl, 2 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2018 Aug 14
|
Structural characterization of the human DjC20/HscB cochaperone in solution
Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics :140970 (2023)
de Souza Coto A, Pereira A, Oliveira S, de Oliveira Moritz M, da Rocha A, Dores-Silva P, da Silva N, de Araújo Nogueira A, Gava L, Seraphim T, Borges J
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDSH7_fit1_model1.png)
|
| Sample: |
MRNA -- proprietary sequence, 400 kDa RNA
(R)-N,N,N-trimethyl-2-3-dioleyloxy-1-propanaminium chloride, 1 kDa lipid
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1 kDa lipid
|
| Buffer: |
10 mM HEPES, 5 mM NaCl, 0.1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Jun 9
|
Quantitative size-resolved characterization of mRNA nanoparticles by in-line coupling of asymmetrical-flow field-flow fractionation with small angle X-ray scattering.
Sci Rep 13(1):15764 (2023)
Graewert MA, Wilhelmy C, Bacic T, Schumacher J, Blanchet C, Meier F, Drexel R, Welz R, Kolb B, Bartels K, Nawroth T, Klein T, Svergun D, Langguth P, Haas H
|
|
|
|
|
|
|

|
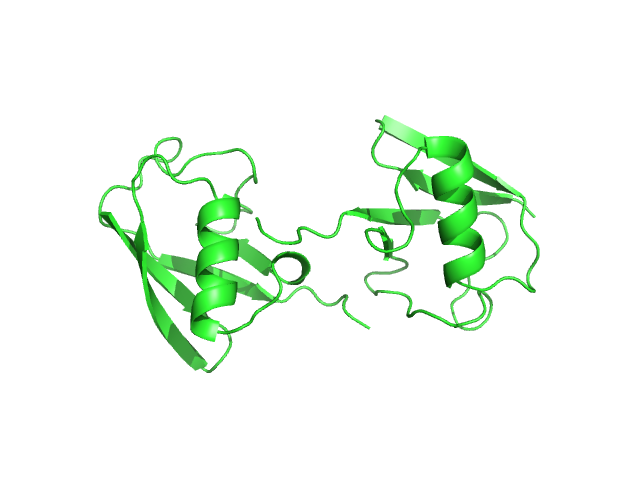
| Sample: |
De novo enterobactin esterase Syn-F4 (K4T) dimer, 25 kDa synthetic construct protein
|
| Buffer: |
25 mM MES, 100 mM NaCl, 10% glycerol, 200 mM Arg-HCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2018 Feb 11
|
Crystal structure and activity of a de novo enzyme, ferric enterobactin esterase Syn-F4.
Proc Natl Acad Sci U S A 120(38):e2218281120 (2023)
Kurihara K, Umezawa K, Donnelly AE, Sperling B, Liao G, Hecht MH, Arai R
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|
|
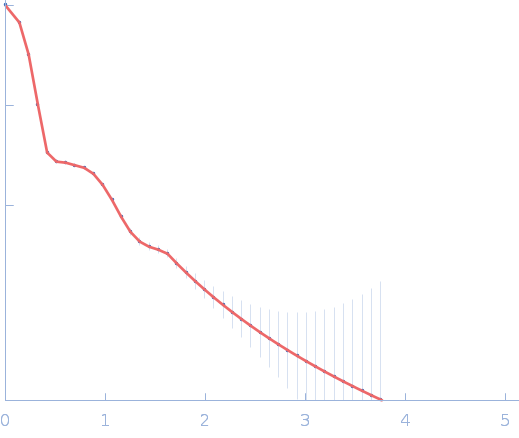
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component tetramer, 256 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (E121C, C162T, C552S, N556C) octamer, 566 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS, Spallation Neutron Source on 2023 Apr 24
|
Domain crossover in the reductase subunit of NADPH-dependent assimilatory sulfite reductase.
J Struct Biol 215(4):108028 (2023)
Walia N, Murray DT, Garg Y, He H, Weiss KL, Nagy G, Elizabeth Stroupe M
|
| RgGuinier |
7.5 |
nm |
| Dmax |
22.6 |
nm |
| VolumePorod |
823 |
nm3 |
|
|
|
|
|
|

|
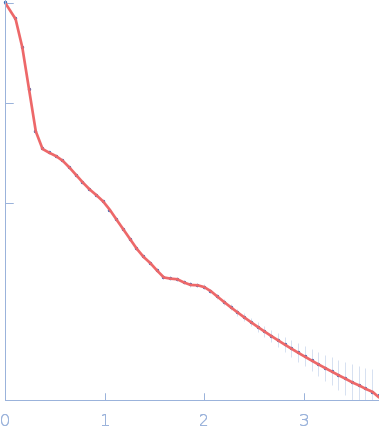
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component tetramer, 256 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (E121C, C162T, C552S, N556C) octamer, 566 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS, Spallation Neutron Source on 2022 Aug 24
|
Domain crossover in the reductase subunit of NADPH-dependent assimilatory sulfite reductase.
J Struct Biol 215(4):108028 (2023)
Walia N, Murray DT, Garg Y, He H, Weiss KL, Nagy G, Elizabeth Stroupe M
|
| RgGuinier |
11.1 |
nm |
| Dmax |
32.4 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component tetramer, 256 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (E121C, C162T, C552S, N556C) octamer, 566 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS, Spallation Neutron Source on 2022 Jul 15
|
Domain crossover in the reductase subunit of NADPH-dependent assimilatory sulfite reductase.
J Struct Biol 215(4):108028 (2023)
Walia N, Murray DT, Garg Y, He H, Weiss KL, Nagy G, Elizabeth Stroupe M
|
| RgGuinier |
10.3 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
1170 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Polyubiquitin-C monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 1
|
Stretching the chains: the destabilizing impact of Cu(2+) and Zn(2+) ions on K48-linked diubiquitin.
Dalton Trans (2023)
Mangini V, Grasso G, Belviso BD, Sciacca MFM, Lanza V, Caliandro R, Milardi D
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
22 |
nm3 |
|
|