|
|
|
|
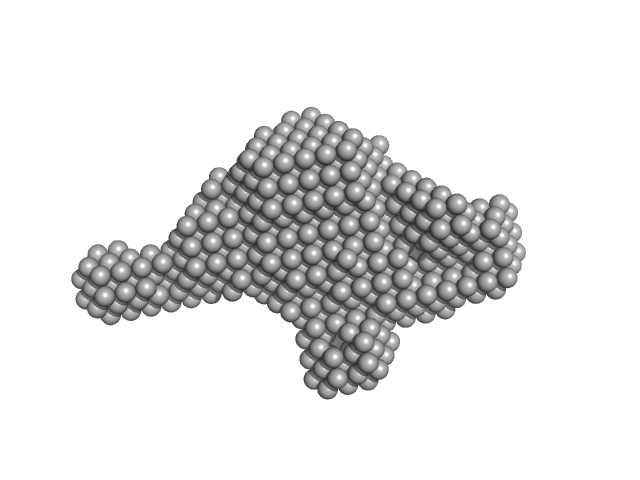
|
| Sample: |
Ras GTPase-activating protein 1 monomer, 101 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8 350 mM NaCl 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Dec 12
|
Tandem engagement of phosphotyrosines by the dual SH2 domains of p120RasGAP.
Structure (2022)
Stiegler AL, Vish KJ, Boggon TJ
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
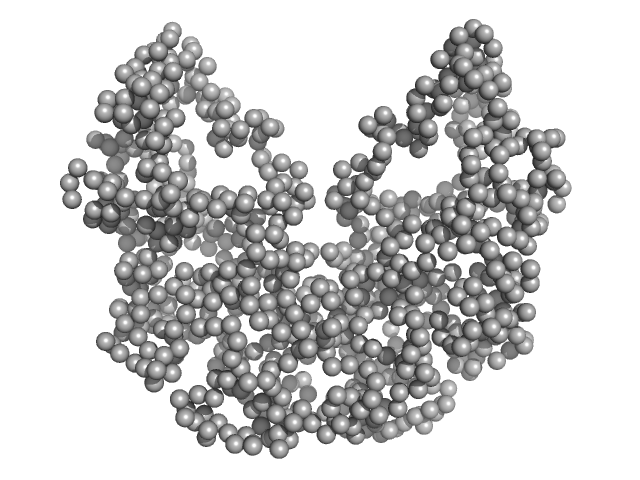
|
| Sample: |
Beta sliding clamp dimer, 86 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris pH 8.0, 200 mM NaCl , 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 15
|
Regulation of futile ligation during early steps of BER in M. tuberculosis is carried out by a β-clamp-XthA-LigA tri-component complex
International Journal of Biological Macromolecules (2022)
Shukla A, Afsar M, Khanam T, Kumar N, Ali F, Kumar S, Jahan F, Ramachandran R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Beta sliding clamp dimer, 86 kDa Mycobacterium tuberculosis protein
Probable exodeoxyribonuclease III protein XthA (Exonuclease III) (EXO III) (AP endonuclease VI) monomer, 33 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 15
|
Regulation of futile ligation during early steps of BER in M. tuberculosis is carried out by a β-clamp-XthA-LigA tri-component complex
International Journal of Biological Macromolecules (2022)
Shukla A, Afsar M, Khanam T, Kumar N, Ali F, Kumar S, Jahan F, Ramachandran R
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
98 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Beta sliding clamp dimer, 86 kDa Mycobacterium tuberculosis protein
Probable exodeoxyribonuclease III protein XthA monomer, 31 kDa Mycobacterium tuberculosis protein
DNA ligase A monomer, 76 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 17
|
Regulation of futile ligation during early steps of BER in M. tuberculosis is carried out by a β-clamp-XthA-LigA tri-component complex
International Journal of Biological Macromolecules (2022)
Shukla A, Afsar M, Khanam T, Kumar N, Ali F, Kumar S, Jahan F, Ramachandran R
|
| RgGuinier |
5.8 |
nm |
| Dmax |
25.3 |
nm |
| VolumePorod |
501 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Probable exodeoxyribonuclease III protein XthA monomer, 31 kDa Mycobacterium tuberculosis protein
DNA ligase A monomer, 76 kDa Mycobacterium tuberculosis protein
Beta sliding clamp dimer, 86 kDa Mycobacterium tuberculosis protein
DNA ligase A nicked DNA substrate dimer, 16 kDa DNA
|
| Buffer: |
50 mM Tris pH 8.0, 200 mM NaCl , 2 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 May 17
|
Regulation of futile ligation during early steps of BER in M. tuberculosis is carried out by a β-clamp-XthA-LigA tri-component complex
International Journal of Biological Macromolecules (2022)
Shukla A, Afsar M, Khanam T, Kumar N, Ali F, Kumar S, Jahan F, Ramachandran R
|
| RgGuinier |
5.8 |
nm |
| Dmax |
19.1 |
nm |
| VolumePorod |
496 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Conjugal transfer mating pair stabilization protein TraG monomer, 52 kDa Shigella flexneri 4c protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5% glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 23
|
Solution characterization of the dynamic conjugative entry exclusion protein TraG
Structural Dynamics 9(6):064702 (2022)
Bragagnolo N, Audette G
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Conjugal transfer mating pair stabilization protein TraG monomer, 57 kDa Shigella flexneri 4c protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5% glycerol, 0.05% NP40, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 23
|
Solution characterization of the dynamic conjugative entry exclusion protein TraG
Structural Dynamics 9(6):064702 (2022)
Bragagnolo N, Audette G
|
| RgGuinier |
5.8 |
nm |
| Dmax |
45.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Phosphoprotein tetramer, 47 kDa Borna disease virus … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM β- mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Sep 1
|
Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses 14(11) (2022)
Tarbouriech N, Chenavier F, Kawasaki J, Bachiri K, Bourhis JM, Legrand P, Freslon LL, Laurent EMN, Suberbielle E, Ruigrok RWH, Tomonaga K, Gonzalez-Dunia D, Horie M, Coyaud E, Crépin T
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
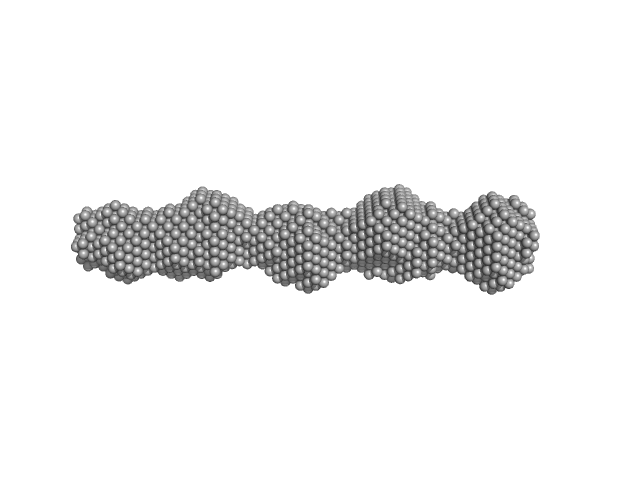
|
| Sample: |
Phosphoprotein oligomerisation domain tetramer, 50 kDa protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM β- mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Sep 1
|
Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses 14(11) (2022)
Tarbouriech N, Chenavier F, Kawasaki J, Bachiri K, Bourhis JM, Legrand P, Freslon LL, Laurent EMN, Suberbielle E, Ruigrok RWH, Tomonaga K, Gonzalez-Dunia D, Horie M, Coyaud E, Crépin T
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Phosphoprotein tetramer, 52 kDa Gaboon viper virus … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM β- mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Sep 1
|
Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses 14(11) (2022)
Tarbouriech N, Chenavier F, Kawasaki J, Bachiri K, Bourhis JM, Legrand P, Freslon LL, Laurent EMN, Suberbielle E, Ruigrok RWH, Tomonaga K, Gonzalez-Dunia D, Horie M, Coyaud E, Crépin T
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
75 |
nm3 |
|
|