|
|
|
|
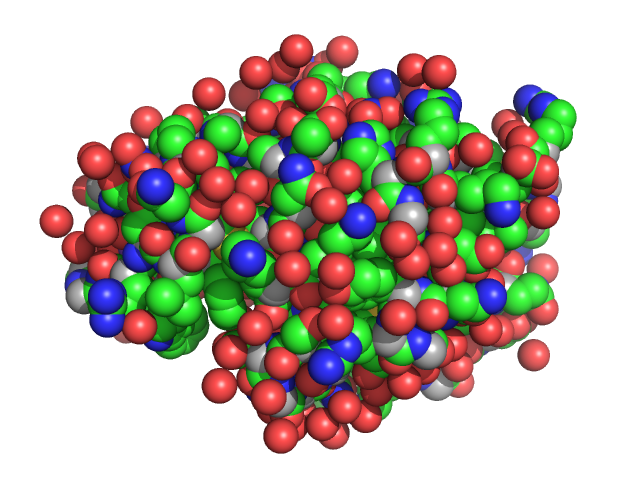
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
200 mM K/Na tartrate, 100 mM tri-sodium citrate pH 5.6, 2.0 M ammonium sulfate, pH: 5.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 28
|
The Relationship of Precursor Cluster Concentration in a Saturated Crystallization Solution to Long-Range Order During the Transition to the Solid Phase.
Acta Naturae 15(1):58-68 (2023)
Marchenkova MA, Boikova AS, Ilina KB, Konarev PV, Pisarevsky YV, Dyakova YA, Kovalchuk MV
|
|
|
|
|
|
|

|
| Sample: |
Dihydroneopterin aldolase tetramer, 55 kDa Helicobacter pylori (strain … protein
|
| Buffer: |
25 mM Tris-HCl pH 7.5 and 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Apr 5
|
Structure of Helicobacter pylori dihydroneopterin aldolase suggests a fragment-based strategy for isozyme-specific inhibitor design.
Curr Res Struct Biol 5:100095 (2023)
Shaw GX, Fan L, Cherry S, Shi G, Tropea JE, Ji X
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
TIP60 (K67E) mutant (metal-ion induced 60-mer complex with barium ions), 1066 kDa Artificial protein protein
Barium ion 0, 8 kDa
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 5% glycerol, 5 mM BaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 May 23
|
Reversible Assembly of an Artificial Protein Nanocage Using Alkaline Earth Metal Ions.
J Am Chem Soc (2022)
Ohara N, Kawakami N, Arai R, Adachi N, Moriya T, Kawasaki M, Miyamoto K
|
| RgGuinier |
9.6 |
nm |
| Dmax |
21.8 |
nm |
|
|
|
|
|
|

|
| Sample: |
TIP60 (K67E) mutant with EDTA dimer, 36 kDa Artificial protein protein
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 1 mM EDTA, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 May 23
|
Reversible Assembly of an Artificial Protein Nanocage Using Alkaline Earth Metal Ions.
J Am Chem Soc (2022)
Ohara N, Kawakami N, Arai R, Adachi N, Moriya T, Kawasaki M, Miyamoto K
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Replicase polyprotein 1ab monomer, 31 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 500 mM NaCl, 5% glycerol, and 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Oct 15
|
Biochemical and structural insights into SARS-CoV-2 polyprotein processing by Mpro.
Sci Adv 8(49):eadd2191 (2022)
Yadav R, Courouble VV, Dey SK, Harrison JJEK, Timm J, Hopkins JB, Slack RL, Sarafianos SG, Ruiz FX, Griffin PR, Arnold E
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Replicase polyprotein 1a monomer, 60 kDa Severe acute respiratory … protein
|
| Buffer: |
20 mM HEPES, 10% glycerol, 500 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 14
|
Biochemical and structural insights into SARS-CoV-2 polyprotein processing by Mpro.
Sci Adv 8(49):eadd2191 (2022)
Yadav R, Courouble VV, Dey SK, Harrison JJEK, Timm J, Hopkins JB, Slack RL, Sarafianos SG, Ruiz FX, Griffin PR, Arnold E
|
| RgGuinier |
3.5 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Multidrug resistance operon repressor dimer, 32 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 10mM DTT, 1% v/v glycerol, pH: 7.1 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Nov 23
|
Small-angle X-ray and neutron scattering of MexR and its complex with DNA supports a conformational selection binding model
Biophysical Journal (2022)
Caporaletti F, Pietras Z, Morad V, Mårtensson L, Gabel F, Wallner B, Martel A, Sunnerhagen M
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
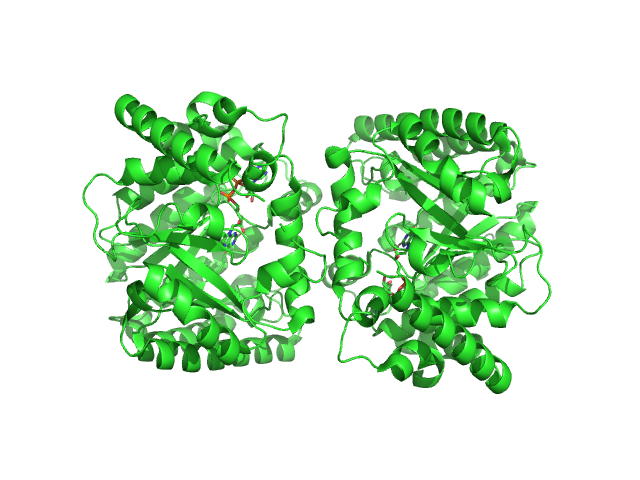
|
| Sample: |
Minimal proline dehydrogenase domain of proline utilization A (design #2) dimer, 87 kDa Sinorhizobium meliloti protein
|
| Buffer: |
25 mM HEPES pH 7.6, 150 mM NaCl, and 1mM TCEP, pH: 7.6 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 12
|
Structure-based engineering of minimal Proline dehydrogenase domains for inhibitor discovery.
Protein Eng Des Sel (2022)
Bogner AN, Ji J, Tanner JJ
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
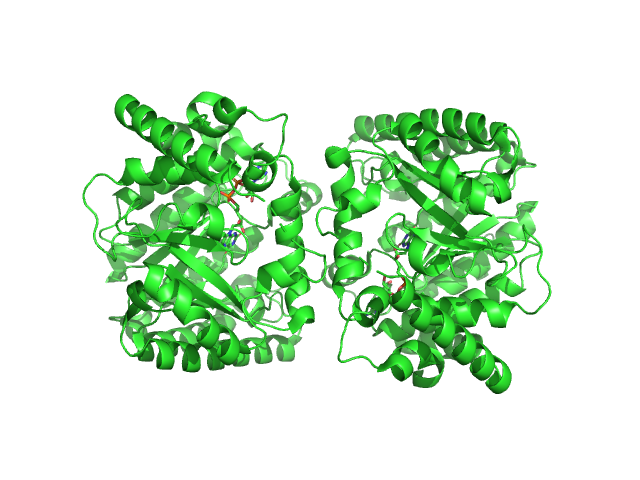
|
| Sample: |
Minimal proline dehydrogenase domain of proline utilization A (design #2) dimer, 87 kDa Sinorhizobium meliloti protein
|
| Buffer: |
25 mM HEPES pH 7.6, 150 mM NaCl, and 1mM TCEP, pH: 7.6 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 12
|
Structure-based engineering of minimal Proline dehydrogenase domains for inhibitor discovery.
Protein Eng Des Sel (2022)
Bogner AN, Ji J, Tanner JJ
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
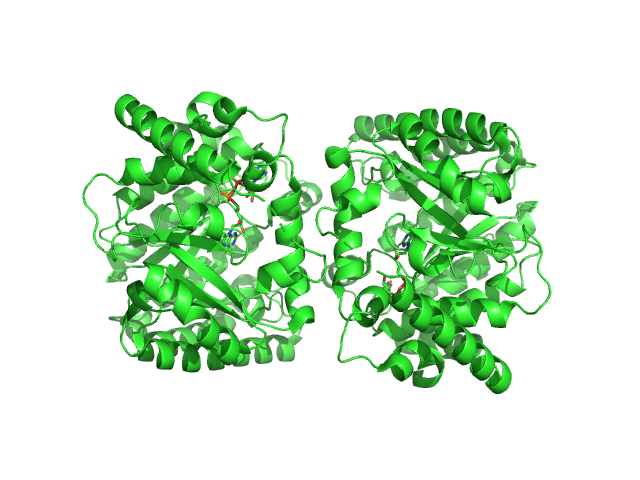
|
| Sample: |
Minimal proline dehydrogenase domain of proline utilization A (design #2) dimer, 87 kDa Sinorhizobium meliloti protein
|
| Buffer: |
25 mM HEPES pH 7.6, 150 mM NaCl, and 1mM TCEP, pH: 7.6 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 12
|
Structure-based engineering of minimal Proline dehydrogenase domains for inhibitor discovery.
Protein Eng Des Sel (2022)
Bogner AN, Ji J, Tanner JJ
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
108 |
nm3 |
|
|