|
|
|
|

|
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 37 kDa Homo sapiens protein
Protein phosphatase 1 regulatory subunit 3C monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 0.5 M NaCl, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 7
|
Molecular architecture of the glycogen- committed PP1/PTG holoenzyme.
Nat Commun 13(1):6199 (2022)
Semrau MS, Giachin G, Covaceuszach S, Cassetta A, Demitri N, Storici P, Lolli G
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Serine/threonine-protein phosphatase PP1-alpha catalytic subunit monomer, 37 kDa Homo sapiens protein
Protein phosphatase 1 regulatory subunit 3C monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 0.5 M NaCl, 10% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 7
|
Molecular architecture of the glycogen- committed PP1/PTG holoenzyme.
Nat Commun 13(1):6199 (2022)
Semrau MS, Giachin G, Covaceuszach S, Cassetta A, Demitri N, Storici P, Lolli G
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
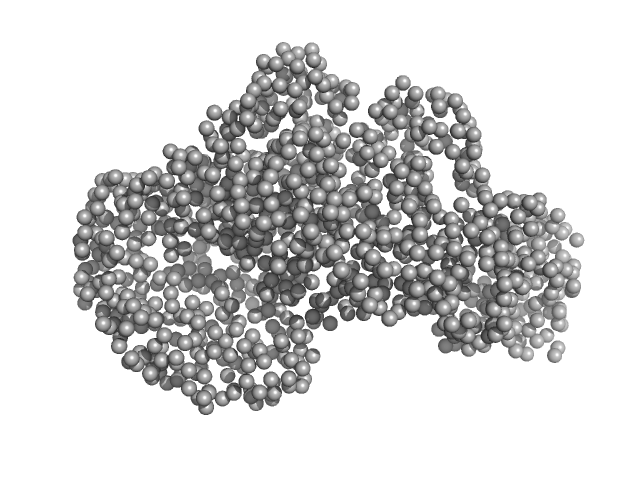
|
| Sample: |
Piwi domain protein monomer, 53 kDa Geobacter sulfurreducens (strain … protein
Sir2 superfamily protein monomer, 68 kDa Geobacter sulfurreducens (strain … protein
|
| Buffer: |
20 mM TrisHCl, pH 7.5, 5 mM MgCl2, 150 mM NaCl and 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 27
|
Short prokaryotic Argonautes provide defence against incoming mobile genetic elements through NAD+ depletion
Nature Microbiology (2022)
Zaremba M, Dakineviciene D, Golovinas E, Zagorskaitė E, Stankunas E, Lopatina A, Sorek R, Manakova E, Ruksenaite A, Silanskas A, Asmontas S, Grybauskas A, Tylenyte U, Jurgelaitis E, Grigaitis R, Timinskas K, Venclovas Č, Siksnys V
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
185 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Elongation factor Tu monomer, 44 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Jul 12
|
Structural insights of the elongation factor EF-Tu complexes in protein translation of Mycobacterium tuberculosis.
Commun Biol 5(1):1052 (2022)
Zhan B, Gao Y, Gao W, Li Y, Li Z, Qi Q, Lan X, Shen H, Gan J, Zhao G, Li J
|
| RgGuinier |
3.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Elongation factor Ts monomer, 29 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Jul 12
|
Structural insights of the elongation factor EF-Tu complexes in protein translation of Mycobacterium tuberculosis.
Commun Biol 5(1):1052 (2022)
Zhan B, Gao Y, Gao W, Li Y, Li Z, Qi Q, Lan X, Shen H, Gan J, Zhao G, Li J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Elongation factor Tu monomer, 44 kDa Mycobacterium tuberculosis (strain … protein
Elongation factor Ts monomer, 29 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Jul 12
|
Structural insights of the elongation factor EF-Tu complexes in protein translation of Mycobacterium tuberculosis.
Commun Biol 5(1):1052 (2022)
Zhan B, Gao Y, Gao W, Li Y, Li Z, Qi Q, Lan X, Shen H, Gan J, Zhao G, Li J
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Chromatin assembly factor 1 subunit A monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 200 mM NaCl, 0.1 mM TCEP, pH: 7 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2021 Oct 28
|
Unorthodox PCNA Binding by Chromatin Assembly Factor 1
International Journal of Molecular Sciences 23(19):11099 (2022)
Gopinathan Nair A, Rabas N, Lejon S, Homiski C, Osborne M, Cyr N, Sverzhinsky A, Melendy T, Pascal J, Laue E, Borden K, Omichinski J, Verreault A
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
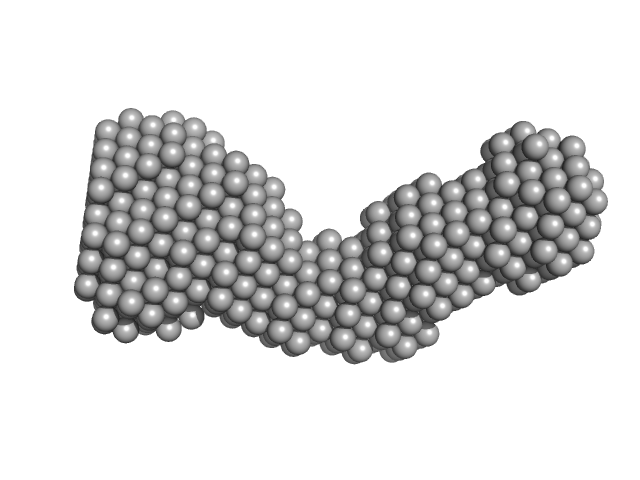
|
| Sample: |
Sperm acrosome membrane-associated protein 6 monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
2.7 mM KCl, 137 mM NaCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH: 7.4 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2021 Dec 11
|
SPACA6 ectodomain structure reveals a conserved superfamily of gamete fusion-associated proteins.
Commun Biol 5(1):984 (2022)
Vance TDR, Yip P, Jiménez E, Li S, Gawol D, Byrnes J, Usón I, Ziyyat A, Lee JE
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
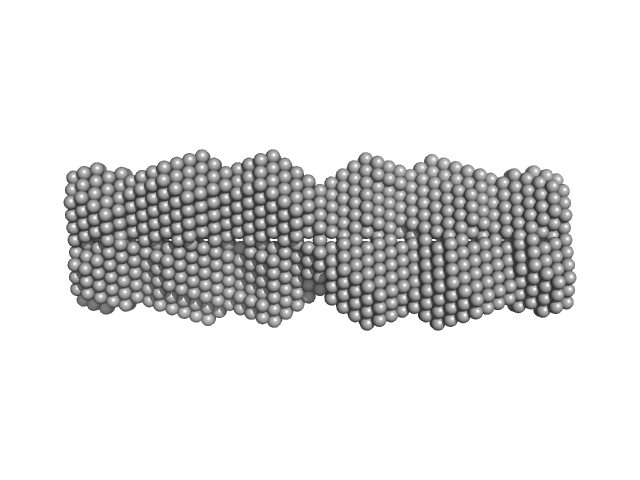
|
| Sample: |
Testis-expressed protein 12 (L110E, F114E, I117E, L121E) tetramer, 36 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Sep 15
|
Coiled-coil structure of meiosis protein TEX12 and conformational regulation by its C-terminal tip
Communications Biology 5(1) (2022)
Dunce J, Salmon L, Davies O
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
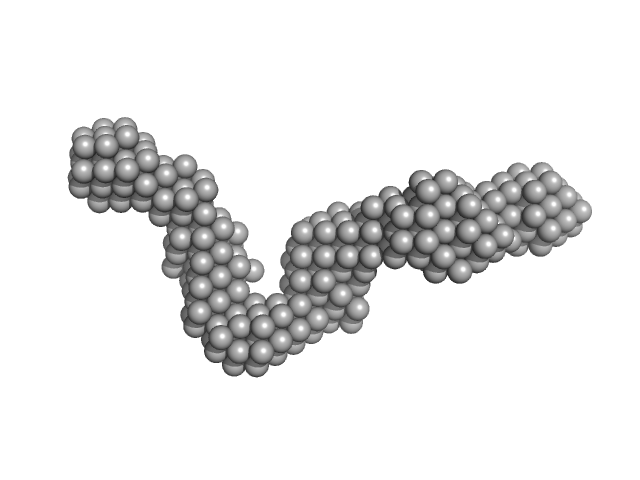
|
| Sample: |
40-mer single stranded inhibitory DNA monomer, 12 kDa DNA
|
| Buffer: |
50 mM phosphate pH 6.0, 200 mM NaCl, 2 mM β-mercaptoethanol (β-ME), 5% glycerol, 200 µM Na2-EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 8
|
Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity
Viruses 14(9):1974 (2022)
Barzak F, Ryan T, Mohammadzadeh N, Harjes S, Kvach M, Kurup H, Krause K, Chelico L, Filichev V, Harjes E, Jameson G
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
26 |
nm3 |
|
|