|
|
|
|

|
| Sample: |
DNA dC->dU-editing enzyme APOBEC-3G tetramer, 186 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate pH 6.0, 200 mM NaCl, 2 mM β-mercaptoethanol (β-ME), 5% glycerol, 200 µM Na2-EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 6
|
Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity
Viruses 14(9):1974 (2022)
Barzak F, Ryan T, Mohammadzadeh N, Harjes S, Kvach M, Kurup H, Krause K, Chelico L, Filichev V, Harjes E, Jameson G
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
350 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA dC->dU-editing enzyme APOBEC-3G tetramer, 186 kDa Homo sapiens protein
40-mer single stranded inhibitory DNA dimer, 24 kDa DNA
|
| Buffer: |
50 mM phosphate pH 6.0, 200 mM NaCl, 2 mM β-mercaptoethanol (β-ME), 5% glycerol, 200 µM Na2-EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 6
|
Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity
Viruses 14(9):1974 (2022)
Barzak F, Ryan T, Mohammadzadeh N, Harjes S, Kvach M, Kurup H, Krause K, Chelico L, Filichev V, Harjes E, Jameson G
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
395 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
40-mer single stranded inhibitory DNA monomer, 12 kDa DNA
DNA dC->dU-editing enzyme APOBEC-3G monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM phosphate pH 6.0, 200 mM NaCl, 2 mM β-mercaptoethanol (β-ME), 5% glycerol, 200 µM Na2-EDTA, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 6
|
Small-Angle X-ray Scattering (SAXS) Measurements of APOBEC3G Provide Structural Basis for Binding of Single-Stranded DNA and Processivity
Viruses 14(9):1974 (2022)
Barzak F, Ryan T, Mohammadzadeh N, Harjes S, Kvach M, Kurup H, Krause K, Chelico L, Filichev V, Harjes E, Jameson G
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
118 |
nm3 |
|
|
|
|
|
|
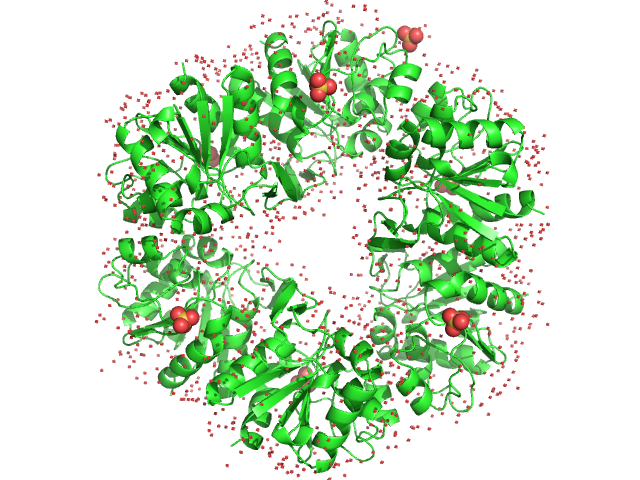
|
| Sample: |
Deglycase PH1704 hexamer, 112 kDa Pyrococcus horikoshii (strain … protein
|
| Buffer: |
20 mM Tris pH 7.5, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Jun 7
|
Medical contrast agents as promising tools for biomacromolecular SAXS experiments.
Acta Crystallogr D Struct Biol 78(Pt 9):1120-1130 (2022)
Gabel F, Engilberge S, Schmitt E, Thureau A, Mechulam Y, Pérez J, Girard E
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|
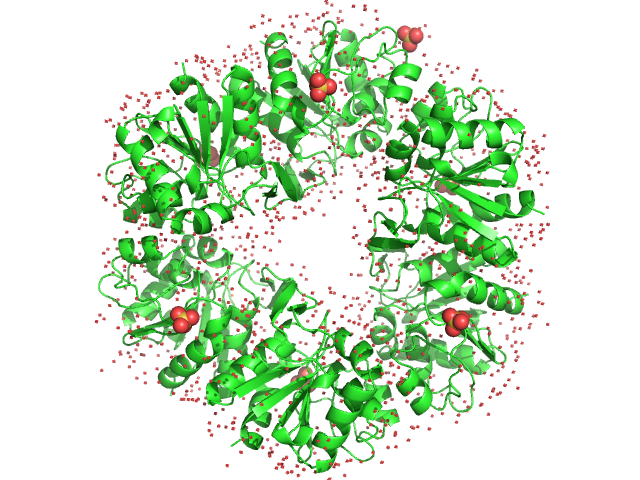
|
| Sample: |
Deglycase PH1704 hexamer, 112 kDa Pyrococcus horikoshii (strain … protein
|
| Buffer: |
20 mM Tris pH 7.5 and 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Nov 26
|
Medical contrast agents as promising tools for biomacromolecular SAXS experiments.
Acta Crystallogr D Struct Biol 78(Pt 9):1120-1130 (2022)
Gabel F, Engilberge S, Schmitt E, Thureau A, Mechulam Y, Pérez J, Girard E
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
140 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
C-phycocyanin alpha subunit trimer, 53 kDa Arthrospira platensis protein
C-phycocyanin beta subunit trimer, 54 kDa Arthrospira platensis protein
|
| Buffer: |
150 mM NaCl, 20 mM Tris, pH: 7.6 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2022 Apr 14
|
Anti-Stokes fluorescence excitation reveals conformational mobility of the C-phycocyanin chromophores
Structural Dynamics 9(5):054701 (2022)
Tsoraev G, Protasova E, Klimanova E, Ryzhykau Y, Kuklin A, Semenov Y, Ge B, Li W, Qin S, Friedrich T, Sluchanko N, Maksimov E
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
154 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
C-phycocyanin alpha subunit monomer, 18 kDa Arthrospira platensis protein
C-phycocyanin beta subunit monomer, 18 kDa Arthrospira platensis protein
|
| Buffer: |
8 M urea, 30 mM βME, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2022 Apr 14
|
Anti-Stokes fluorescence excitation reveals conformational mobility of the C-phycocyanin chromophores
Structural Dynamics 9(5):054701 (2022)
Tsoraev G, Protasova E, Klimanova E, Ryzhykau Y, Kuklin A, Semenov Y, Ge B, Li W, Qin S, Friedrich T, Sluchanko N, Maksimov E
|
| RgGuinier |
6.6 |
nm |
| Dmax |
23.5 |
nm |
|
|
|
|
|
|
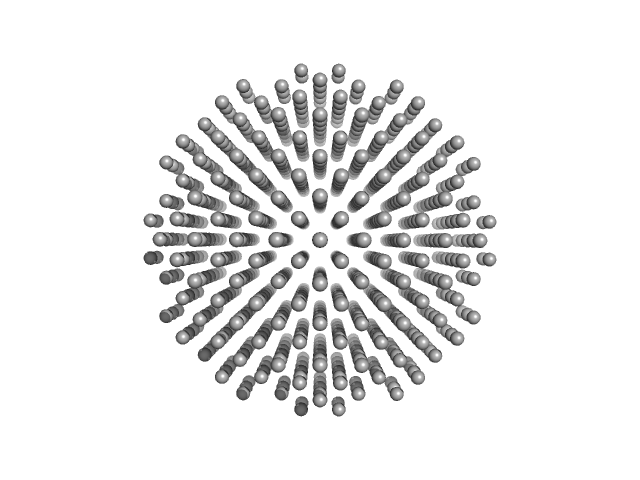
|
| Sample: |
Nominal 100 nm diameter polystyrene spheres monomer, 315312 kDa
|
| Buffer: |
Water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 May 16
|
Standard polystyrene nanospheres (concentration series data)
Dima Molodenskiy
|
| RgGuinier |
33.0 |
nm |
| Dmax |
100.0 |
nm |
| VolumePorod |
520 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Nominal 20 nm diameter polystyrene spheres 0, 8 kDa
|
| Buffer: |
Water, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 May 16
|
Standard polystyrene nanospheres (concentration series data)
Dima Molodenskiy
|
| RgGuinier |
8.5 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
|
|
|
|
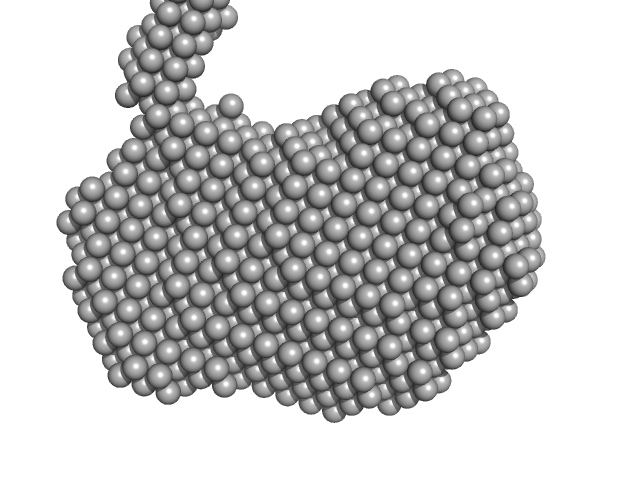
|
| Sample: |
LipAMS8 monomer, 50 kDa Pseudomonas sp. AMS8 protein
|
| Buffer: |
50 mM Tris HCl, 5 mM CaCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2018 Mar 6
|
Structural interpretations of a flexible cold-active AMS8 lipase by combining small-angle X-ray scattering and molecular dynamics simulation (SAXS-MD).
Int J Biol Macromol (2022)
Yaacob N, Kamonsutthipaijit N, Soontaranon S, Leow TC, Rahman RNZRA, Ali MSM
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
87 |
nm3 |
|
|