|
|
|
|
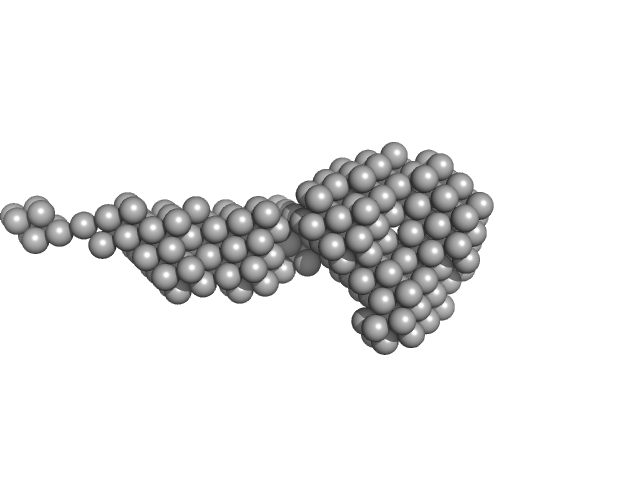
|
| Sample: |
Lectin nano-block WA20-ΔN3ACG dimer, 58 kDa protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5% glycerol,, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Oct 31
|
Self-Assembling Lectin Nano-Block Oligomers Enhance Binding Avidity to Glycans
International Journal of Molecular Sciences 23(2):676 (2022)
Irumagawa S, Hiemori K, Saito S, Tateno H, Arai R
|
| RgGuinier |
4.1 |
nm |
| Dmax |
21.6 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Francisella tularensis outer membrane protein A dimer, 80 kDa Francisella tularensis subsp. … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.05% B-DDM, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Mar 20
|
Structural and biophysical properties of FopA, a major outer membrane protein of Francisella tularensis.
PLoS One 17(8):e0267370 (2022)
Nagaratnam N, Martin-Garcia JM, Yang JH, Goode MR, Ketawala G, Craciunescu FM, Zook JD, Sonowal M, Williams D, Grant TD, Fromme R, Hansen DT, Fromme P
|
| RgGuinier |
4.4 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
330 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Centrosomal-P4.1-associated-protein monomer, 12 kDa Homo sapiens protein
Centriolar coiled-coil protein of 110 kDa dimer, 20 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM DTT, 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 23
|
Centriolar cap proteins CP110 and CPAP control slow elongation of microtubule plus ends.
J Cell Biol 224(3) (2025)
Iyer SS, Chen F, Ogunmolu FE, Moradi S, Volkov VA, van Grinsven EJ, van Hoorn C, Wu J, Andrea N, Hua S, Jiang K, Vakonakis I, Potočnjak M, Herzog F, Gigant B, Gudimchuk N, Stecker KE, Dogterom M, Steinmetz MO, Akhmanova A
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Centriolar coiled-coil protein of 110 kDa dimer, 20 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM DTT, 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Feb 23
|
Centriolar cap proteins CP110 and CPAP control slow elongation of microtubule plus ends.
J Cell Biol 224(3) (2025)
Iyer SS, Chen F, Ogunmolu FE, Moradi S, Volkov VA, van Grinsven EJ, van Hoorn C, Wu J, Andrea N, Hua S, Jiang K, Vakonakis I, Potočnjak M, Herzog F, Gigant B, Gudimchuk N, Stecker KE, Dogterom M, Steinmetz MO, Akhmanova A
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
HomA outer membrane protein dimer, 146 kDa Helicobacter pylori protein
|
| Buffer: |
20 mM Tris-Cl, 200 mM NaCl, 5 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2021 Feb 2
|
Biophysical characterization of the homodimers of HomA and HomB, outer membrane proteins of Helicobacter pylori.
Sci Rep 11(1):24471 (2021)
Tamrakar A, Singh R, Kumar A, Makde RD, Ashish, Kodgire P
|
| RgGuinier |
8.3 |
nm |
| Dmax |
28.2 |
nm |
| VolumePorod |
426 |
nm3 |
|
|
|
|
|
|
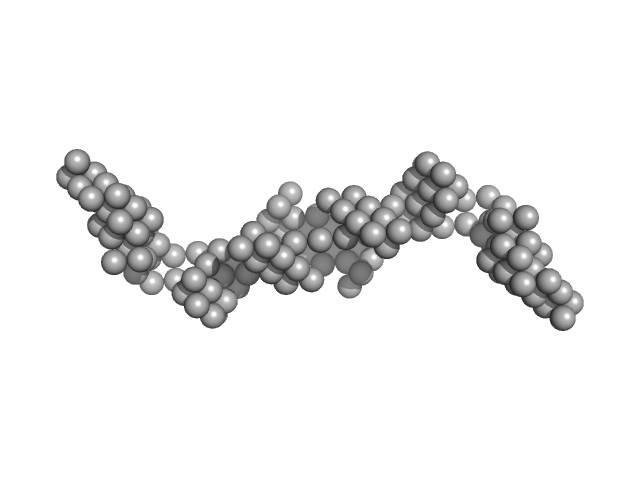
|
| Sample: |
HomB outer membrane protein dimer, 148 kDa Helicobacter pylori protein
|
| Buffer: |
20 mM Tris-Cl, 200 mM NaCl, 5 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2021 Feb 2
|
Biophysical characterization of the homodimers of HomA and HomB, outer membrane proteins of Helicobacter pylori.
Sci Rep 11(1):24471 (2021)
Tamrakar A, Singh R, Kumar A, Makde RD, Ashish, Kodgire P
|
| RgGuinier |
8.2 |
nm |
| Dmax |
27.1 |
nm |
| VolumePorod |
500 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
5-10-5 gapmer phosphorothioate antisense oligonucleotide dimer, 14 kDa
|
| Buffer: |
20 mM Tris-Cl (pH 7.5), 250 mM KCl, 50 mM L-proline, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 28
|
Structural basis of dimerization and nucleic acid binding of human DBHS proteins NONO and PSPC1.
Nucleic Acids Res (2021)
Knott GJ, Chong YS, Passon DM, Liang XH, Deplazes E, Conte MR, Marshall AC, Lee M, Fox AH, Bond CS
|
| RgGuinier |
1.8 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
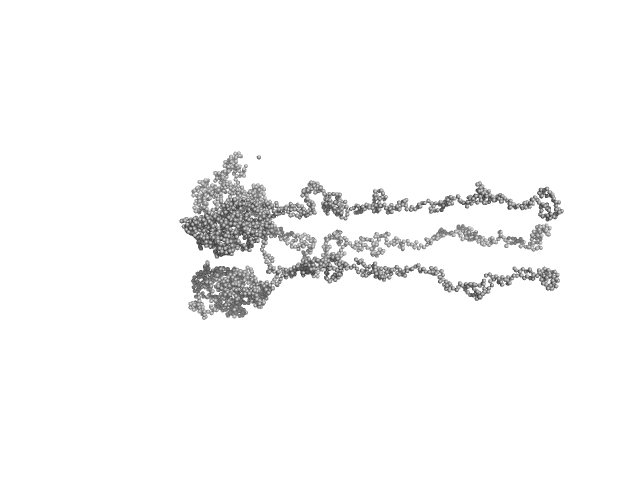
|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
1400 mM NaCl, 49.4 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Feb 10
|
Ambiguities in and completeness of SAS data analysis of membrane proteins: the case of the sensory rhodopsin II–transducer complex
Acta Crystallographica Section D Structural Biology 77(11) (2021)
Ryzhykau Y, Vlasov A, Orekhov P, Rulev M, Rogachev A, Vlasova A, Kazantsev A, Verteletskiy D, Skoi V, Brennich M, Pernot P, Murugova T, Gordeliy V, Kuklin A
|
| RgGuinier |
8.6 |
nm |
| Dmax |
39.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
2800 mM NaCl, 76.6 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Feb 10
|
Ambiguities in and completeness of SAS data analysis of membrane proteins: the case of the sensory rhodopsin II–transducer complex
Acta Crystallographica Section D Structural Biology 77(11) (2021)
Ryzhykau Y, Vlasov A, Orekhov P, Rulev M, Rogachev A, Vlasova A, Kazantsev A, Verteletskiy D, Skoi V, Brennich M, Pernot P, Murugova T, Gordeliy V, Kuklin A
|
| RgGuinier |
9.0 |
nm |
| Dmax |
39.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Sensory rhodopsin II from Natronbacterium pharaonis dimer, 53 kDa Natronomonas pharaonis protein
Sensory rhodopsin II transducer from Natronomonas pharaonis dimer, 116 kDa Natronomonas pharaonis protein
|
| Buffer: |
4000 mM NaCl, 100 mM Na/Na-Pi, 1.0 mM EDTA, 0.05% DDM (D2O buffer), pH: 8 |
| Experiment: |
SANS
data collected at YuMO SANS TOF spectrometer, IBR-2, Frank Laboratory of Neutron Physics, Joint Institute for Nuclear Research on 2019 Jan 25
|
Ambiguities in and completeness of SAS data analysis of membrane proteins: the case of the sensory rhodopsin II–transducer complex
Acta Crystallographica Section D Structural Biology 77(11) (2021)
Ryzhykau Y, Vlasov A, Orekhov P, Rulev M, Rogachev A, Vlasova A, Kazantsev A, Verteletskiy D, Skoi V, Brennich M, Pernot P, Murugova T, Gordeliy V, Kuklin A
|
| RgGuinier |
10.4 |
nm |
| Dmax |
39.0 |
nm |
|
|