|
|
|
|
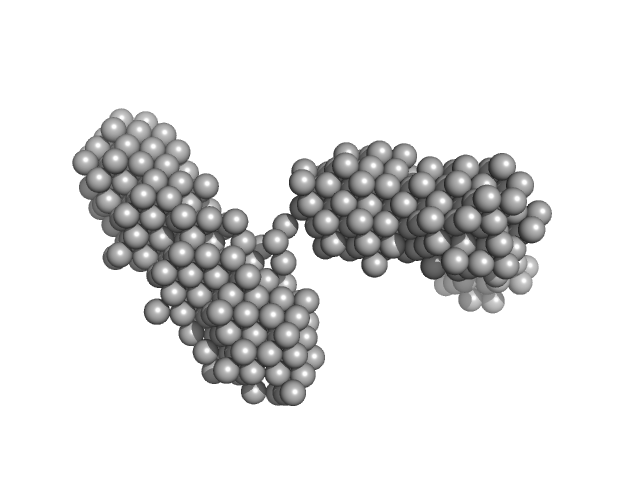
|
| Sample: |
Lysozyme amyloid fibril, 1 kDa Gallus gallus protein
Fe3O4 nanoparticles; nominal diameter 20 nm (hydrodynamic diameter) monomer, 1 kDa
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 29
|
Dependence of the Nanoscale Composite Morphology of Fe3O4 Nanoparticle-Infused Lysozyme Amyloid Fibrils on Timing of Infusion: A Combined SAXS and AFM Study
Molecules 26(16):4864 (2021)
Schroer M, Hu P, Tomasovicova N, Batkova M, Zakutanska K, Wu P, Kopcansky P
|
| RgGuinier |
23.6 |
nm |
| Dmax |
90.0 |
nm |
|
|
|
|
|
|
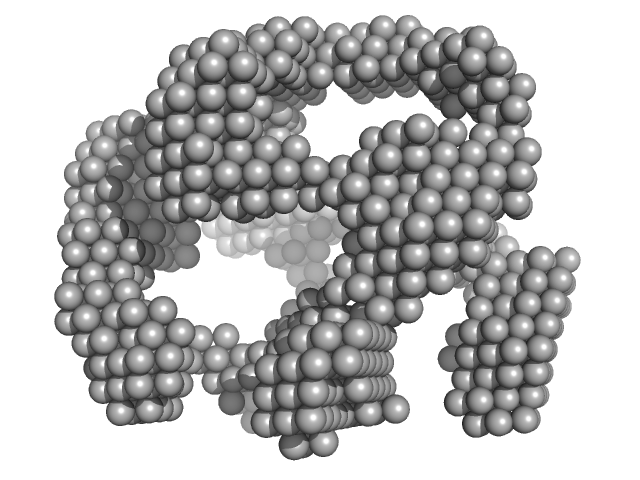
|
| Sample: |
Lysozyme amyloid fibril, 1 kDa Gallus gallus protein
Fe3O4 nanoparticles; nominal diameter 20 nm (hydrodynamic diameter) monomer, 1 kDa
|
| Buffer: |
0.2 M glycine-HCl, 80 mM NaCl, pH: 2.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Sep 2
|
Dependence of the Nanoscale Composite Morphology of Fe3O4 Nanoparticle-Infused Lysozyme Amyloid Fibrils on Timing of Infusion: A Combined SAXS and AFM Study
Molecules 26(16):4864 (2021)
Schroer M, Hu P, Tomasovicova N, Batkova M, Zakutanska K, Wu P, Kopcansky P
|
| RgGuinier |
31.0 |
nm |
| Dmax |
75.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
L-lactate dehydrogenase tetramer, 141 kDa Plasmodium falciparum protein
|
| Buffer: |
100 mM Na-phosphate buffer, 400 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2019 Jul 3
|
A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity
Communications Biology 4(1) (2021)
Reyes Romero A, Lunev S, Popowicz G, Calderone V, Gentili M, Sattler M, Plewka J, Taube M, Kozak M, Holak T, Dömling A, Groves M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
244 |
nm3 |
|
|
|
|
|
|
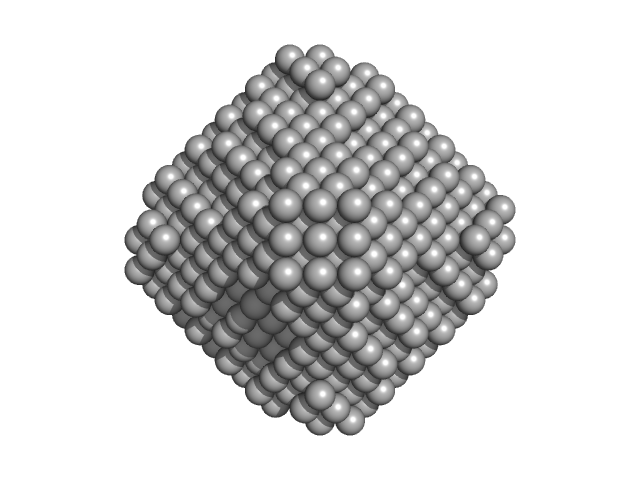
|
| Sample: |
L-lactate dehydrogenase tetramer, 141 kDa Plasmodium falciparum protein
|
| Buffer: |
100 mM Na-phosphate buffer, 400 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 with MetalJet, Department of Macromolecular Physics, Faculty of Physics, Adam Mickiewicz University on 2019 Jul 3
|
A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity
Communications Biology 4(1) (2021)
Reyes Romero A, Lunev S, Popowicz G, Calderone V, Gentili M, Sattler M, Plewka J, Taube M, Kozak M, Holak T, Dömling A, Groves M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
223 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
BON domain protein decamer, 229 kDa Acinetobacter baumannii protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.03 % NaN3, 5.0 % glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Apr 11
|
BonA from Acinetobacter baumannii Forms a Divisome-Localized Decamer That Supports Outer Envelope Function.
mBio :e0148021 (2021)
Grinter R, Morris FC, Dunstan RA, Leung PM, Kropp A, Belousoff M, Gunasinghe SD, Scott NE, Beckham S, Peleg AY, Greening C, Li J, Heinz E, Lithgow T
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
546 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
BON domain protein monomer, 20 kDa Acinetobacter baumannii protein
|
| Buffer: |
20 mM Tris HCl, 150 nM NaCl, 0.02 % NaN3, 5% glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Apr 11
|
BonA from Acinetobacter baumannii Forms a Divisome-Localized Decamer That Supports Outer Envelope Function.
mBio :e0148021 (2021)
Grinter R, Morris FC, Dunstan RA, Leung PM, Kropp A, Belousoff M, Gunasinghe SD, Scott NE, Beckham S, Peleg AY, Greening C, Li J, Heinz E, Lithgow T
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
PSK, an antimicrobial peptide from Chrysomya megacephala monomer, 10 kDa Chrysomya megacephala protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Dec 7
|
Crystal and solution structures of a novel antimicrobial peptide from Chrysomya megacephala.
Acta Crystallogr D Struct Biol 77(Pt 7):894-903 (2021)
Xiao C, Xiao Z, Hu C, Lu J, Cui L, Zhang Y, Dai Y, Zhang Q, Wang S, Liu W
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Myelin P2 disease mutant I50del monomer, 15 kDa protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jul 22
|
Human myelin protein P2: from crystallography to time-lapse membrane imaging and neuropathy-associated variants.
FEBS J (2021)
Uusitalo M, Klenow MB, Laulumaa S, Blakeley MP, Simonsen AC, Ruskamo S, Kursula P
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Human P2 M114T mutant monomer, 15 kDa protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jul 22
|
Human myelin protein P2: from crystallography to time-lapse membrane imaging and neuropathy-associated variants.
FEBS J (2021)
Uusitalo M, Klenow MB, Laulumaa S, Blakeley MP, Simonsen AC, Ruskamo S, Kursula P
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
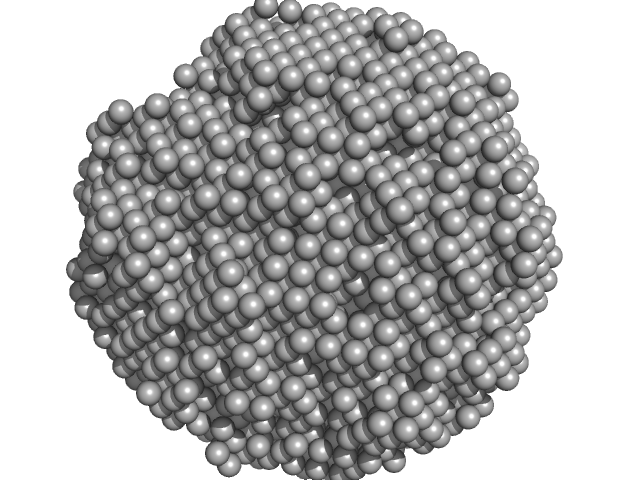
|
| Sample: |
Human P2 V115A mutant monomer, 15 kDa protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jul 22
|
Human myelin protein P2: from crystallography to time-lapse membrane imaging and neuropathy-associated variants.
FEBS J (2021)
Uusitalo M, Klenow MB, Laulumaa S, Blakeley MP, Simonsen AC, Ruskamo S, Kursula P
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
17 |
nm3 |
|
|