|
|
|
|

|
| Sample: |
Collagenase ColH (Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD) with Tyr780Ser, His782Ser, Tyr796Ser and Tyr801Ser) monomer, 23 kDa Hathewaya histolytica protein
|
| Buffer: |
50 mM Tris, 1 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 5
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in V. alginolyticus
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
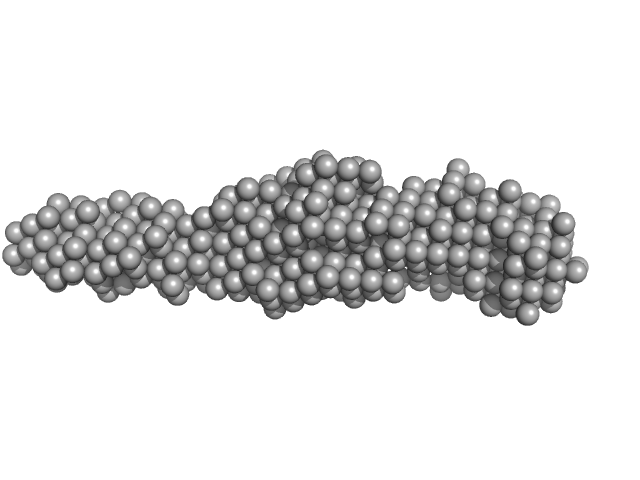
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Collagen like-peptide [GPRG(POG)13] trimer, 11 kDa protein
Collagenase ColH (Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD) with Tyr780Ser, His782Ser, Tyr796Ser and Tyr801Ser) monomer, 23 kDa Hathewaya histolytica protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 5
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in V. alginolyticus
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
| RgGuinier |
3.2 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Collagenase ColH (Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD)) monomer, 23 kDa Hathewaya histolytica protein
Collagen like-peptide [GPRG(POG)13] trimer, 11 kDa protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 5
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in V. alginolyticus
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
| RgGuinier |
3.3 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Pomacea diffusa perivitellin-1, 23 kDa Pomacea diffusa protein
|
| Buffer: |
50 mM Phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2015 Mar 27
|
A highly stable, non-digestible lectin from Pomacea diffusa unveils clade-related protection systems in apple snail eggs.
J Exp Biol 223(Pt 19) (2020)
Brola TR, Dreon MS, Qiu JW, Heras H
|
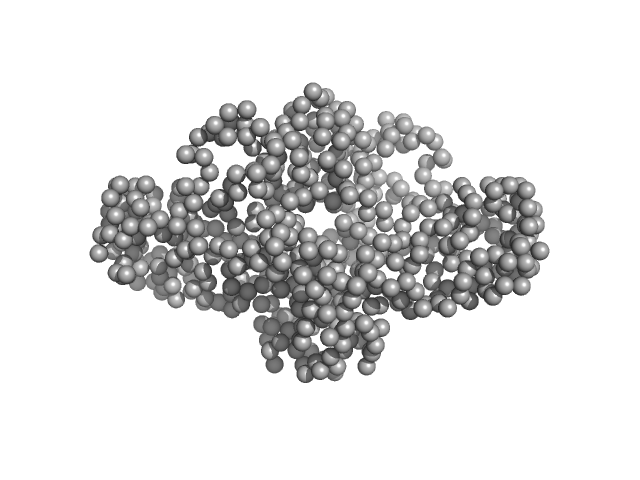
| RgGuinier |
4.6 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
529 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Uncharacterized protein dimer, 66 kDa Legionella pneumophila protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 18
|
Structure, Dynamics and Cellular Insight Into Novel Substrates of the Legionella pneumophila Type II Secretion System
Frontiers in Molecular Biosciences 7 (2020)
Portlock T, Tyson J, Dantu S, Rehman S, White R, McIntire I, Sewell L, Richardson K, Shaw R, Pandini A, Cianciotto N, Garnett J
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
114 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Tegument protein UL7 dimer, 68 kDa Human alphaherpesvirus 1 … protein
Tegument protein UL51 tetramer, 102 kDa Human alphaherpesvirus 1 … protein
|
| Buffer: |
20 mM HEPES, 200 mM NaCl, 3% (v/v) glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 16
|
Insights into herpesvirus assembly from the structure of the pUL7:pUL51 complex.
Elife 9 (2020)
Butt BG, Owen DJ, Jeffries CM, Ivanova L, Hill CH, Houghton JW, Ahmed MF, Antrobus R, Svergun DI, Welch JJ, Crump CM, Graham SC
|
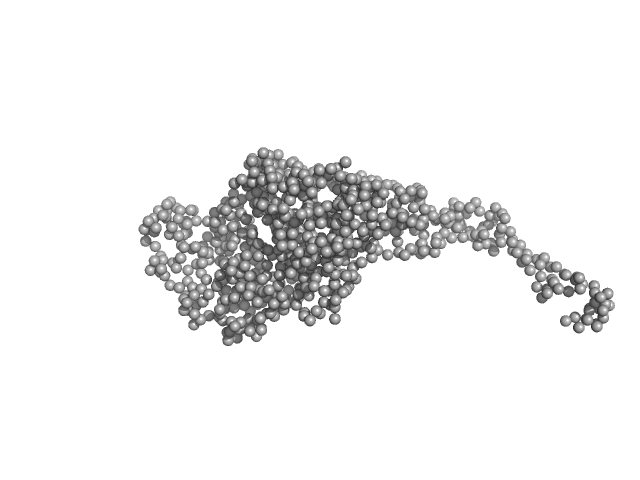
| RgGuinier |
4.6 |
nm |
| Dmax |
19.7 |
nm |
| VolumePorod |
340 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Tegument protein UL7 monomer, 34 kDa Human alphaherpesvirus 1 … protein
Tegument protein UL51 dimer, 51 kDa Human alphaherpesvirus 1 protein
|
| Buffer: |
20 mM HEPES, 200 mM NaCl, 3% (v/v) glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 16
|
Insights into herpesvirus assembly from the structure of the pUL7:pUL51 complex.
Elife 9 (2020)
Butt BG, Owen DJ, Jeffries CM, Ivanova L, Hill CH, Houghton JW, Ahmed MF, Antrobus R, Svergun DI, Welch JJ, Crump CM, Graham SC
|
| RgGuinier |
4.0 |
nm |
| Dmax |
18.2 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
BRME1 dimer, 23 kDa Mus musculus protein
Meiotic localizer of BRCA2 dimer, 26 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 7
|
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells
Nature Communications 11(1) (2020)
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury D, Alam I, Livera G, Emmanuelle M, Wang P, Nandakumar J, Davies O, Shibuya H
|
| RgGuinier |
5.0 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Meiotic localizer of BRCA2 dimer, 26 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 7
|
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells
Nature Communications 11(1) (2020)
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury D, Alam I, Livera G, Emmanuelle M, Wang P, Nandakumar J, Davies O, Shibuya H
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Meiotic localizer of BRCA2 octamer, 102 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 7
|
The BRCA2-MEILB2-BRME1 complex governs meiotic recombination and impairs the mitotic BRCA2-RAD51 function in cancer cells
Nature Communications 11(1) (2020)
Zhang J, Gurusaran M, Fujiwara Y, Zhang K, Echbarthi M, Vorontsov E, Guo R, Pendlebury D, Alam I, Livera G, Emmanuelle M, Wang P, Nandakumar J, Davies O, Shibuya H
|
| RgGuinier |
5.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
368 |
nm3 |
|
|