|
|
|
|

|
| Sample: |
Apolipoprotein A-I dimer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Mar 20
|
The structure of the apolipoprotein A-I monomer provides insights into its oligomerisation and lipid-binding mechanisms
Journal of Molecular Biology :169394 (2025)
Tou H, Rosenes Z, Khandokar Y, Zlatic C, Metcalfe R, Mok Y, Morton C, Gooley P, Griffin M
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
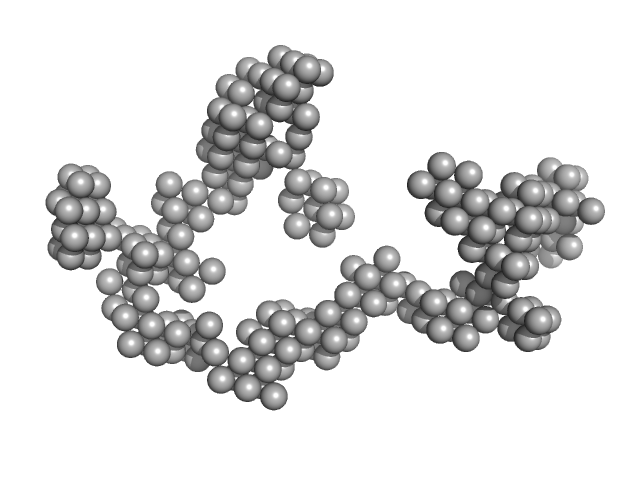
|
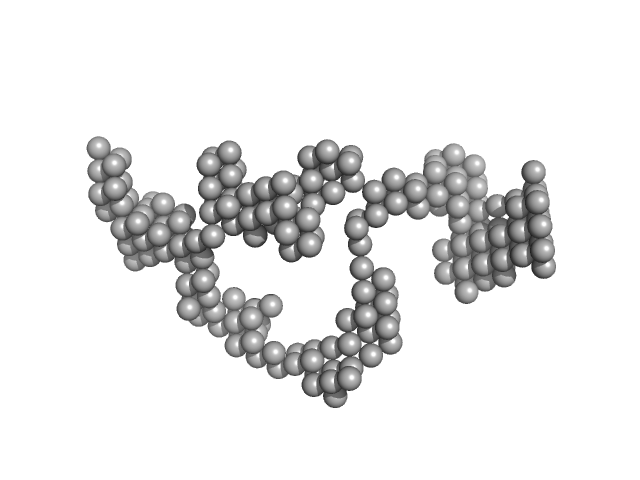
| Sample: |
RNA component of 7SK nuclear ribonucleoprotein monomer, 107 kDa Homo sapiens RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 15 mM KCl, 5% Glycerol, 3 mM MgCl2, pH: 5.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Jun 21
|
Solution Structure Determination and Biophysical Studies of 7SK RNP and 7SL SRP RNAs.
RNA (2025)
D'Souza MH, Pereira HS, Tersteeg S, Vasudeva G, Siddiqui MQ, Agunlejika T, Kerr L, Girodat D, Patel T
|
| RgGuinier |
8.0 |
nm |
| Dmax |
23.2 |
nm |
| VolumePorod |
600 |
nm3 |
|
|
|
|
|
|
|
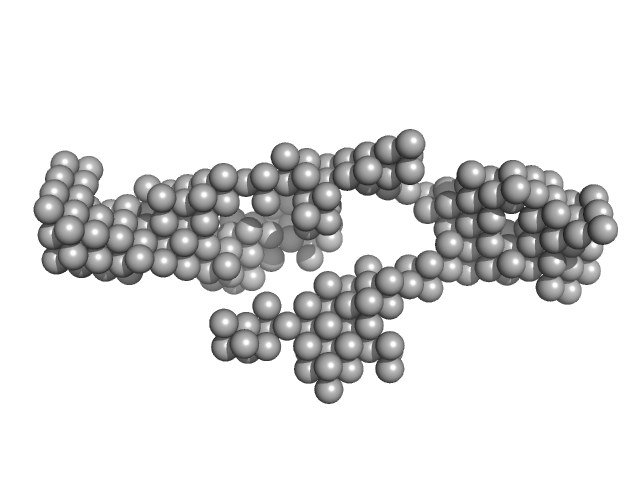
| Sample: |
RNA component of 7SK nuclear ribonucleoprotein monomer, 107 kDa Homo sapiens RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 15 mM KCl, 5% Glycerol, 3 mM MgCl2, pH: 5.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Jun 21
|
Solution Structure Determination and Biophysical Studies of 7SK RNP and 7SL SRP RNAs.
RNA (2025)
D'Souza MH, Pereira HS, Tersteeg S, Vasudeva G, Siddiqui MQ, Agunlejika T, Kerr L, Girodat D, Patel T
|
| RgGuinier |
7.9 |
nm |
| Dmax |
24.1 |
nm |
| VolumePorod |
290 |
nm3 |
|
|
|
|
|
|
|
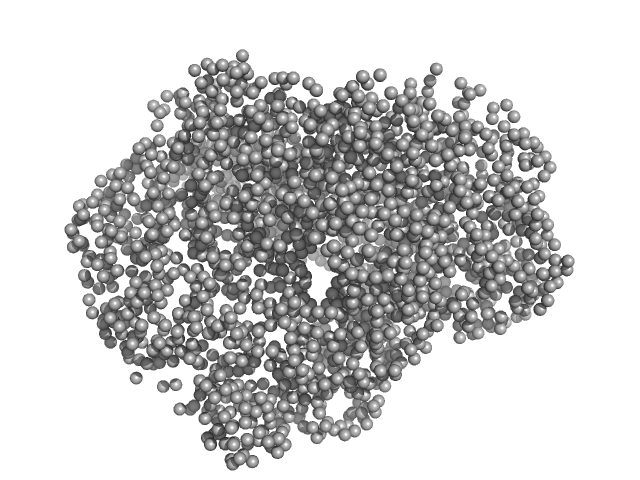
| Sample: |
7SL Signal Reocgnition Particle RNA monomer, 98 kDa Homo sapiens RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 15 mM KCl, 5% Glycerol, 3 mM MgCl2, pH: 5.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Jun 21
|
Solution Structure Determination and Biophysical Studies of 7SK RNP and 7SL SRP RNAs.
RNA (2025)
D'Souza MH, Pereira HS, Tersteeg S, Vasudeva G, Siddiqui MQ, Agunlejika T, Kerr L, Girodat D, Patel T
|
| RgGuinier |
6.7 |
nm |
| Dmax |
21.7 |
nm |
| VolumePorod |
300 |
nm3 |
|
|
|
|
|
|
|
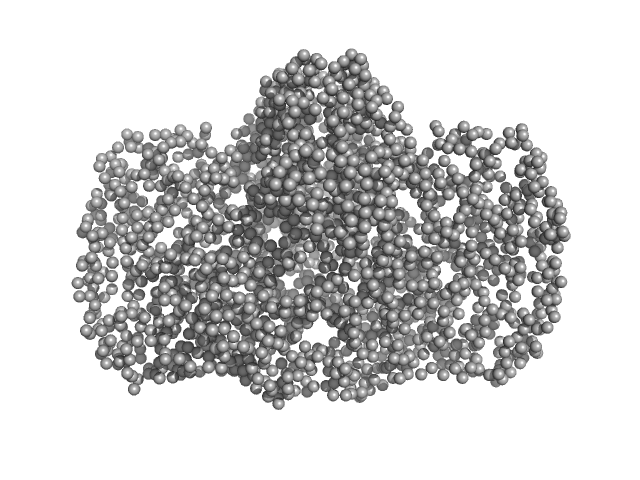
| Sample: |
Thiopeptide-type bacteriocin biosynthesis domain containing protein dimer, 250 kDa Clostridium sp. Maddingley … protein
|
| Buffer: |
50 mM HEPES, 200 mM NaCl, 1 % (v/v) Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 26
|
Structural insights into the substrate binding mechanism of the class I dehydratase MadB
Communications Biology 8(1) (2025)
Knospe C, Ortiz J, Reiners J, Kedrov A, Gertzen C, Smits S, Schmitt L
|
| RgGuinier |
4.5 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
440 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Thiopeptide-type bacteriocin biosynthesis domain containing protein dimer, 250 kDa Clostridium sp. Maddingley … protein
Lantibiotic, gallidermin/nisin family monomer, 3 kDa Clostridium sp. Maddingley … protein
|
| Buffer: |
50 mM HEPES, 200 mM NaCl, 1 % (v/v) Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 3
|
Structural insights into the substrate binding mechanism of the class I dehydratase MadB
Communications Biology 8(1) (2025)
Knospe C, Ortiz J, Reiners J, Kedrov A, Gertzen C, Smits S, Schmitt L
|
| RgGuinier |
4.5 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
445 |
nm3 |
|
|
|
|
|
|

|
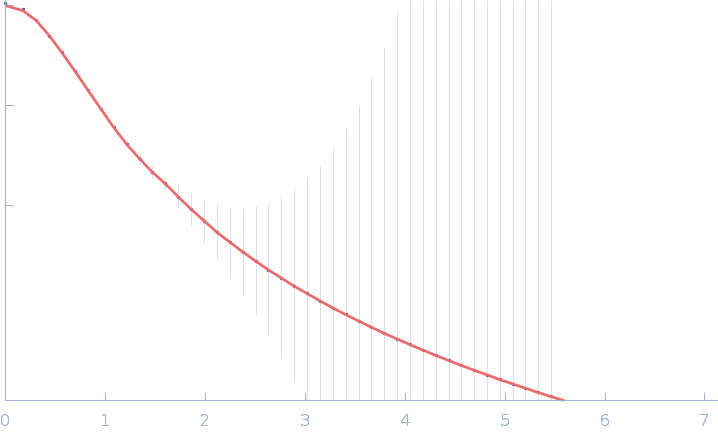
| Sample: |
SH-01 monomer, 41 kDa protein
Fab 3E12 monomer, 50 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jun 15
|
A SELECTIVE ALTERNATIVE PATHWAY COMPLEMENT INHIBITOR FOR TREATMENT OF PAROXYSMAL NOCTURNAL HEMOGLOBINURIA
Hagen Sülzen
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
124 |
nm3 |
|
|
|
|
|
|
|
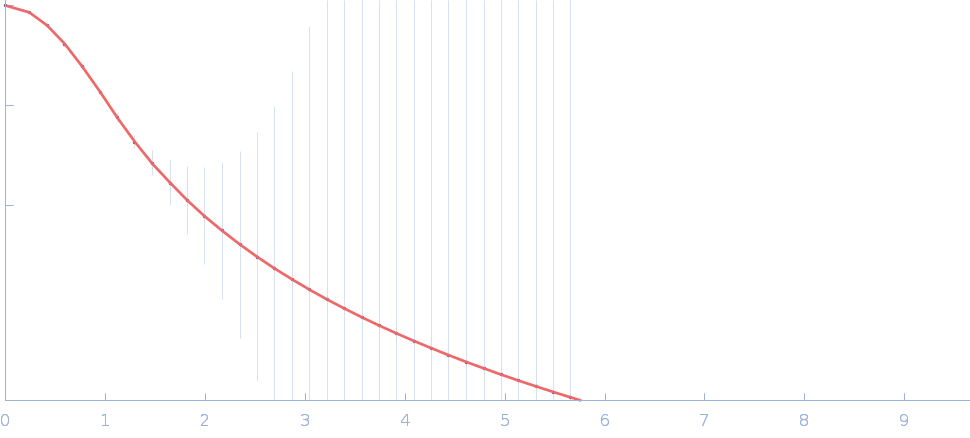
| Sample: |
Fas-activated serine/threonine kinase (amino acids 76-549) monomer, 53 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 5% (v/v) Glycerol, 200 mM Ammonium Sulphate, 50 mM NaCl, 0.5 mM TCEP, 10 mM Glutamic acid, 10 mM Arginine;, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 31
|
Human FASTK preferentially binds single-stranded and G-rich RNA.
FEBS J (2025)
Dawidziak DM, Dzadz DA, Kuska MI, Kanavalli M, Klimecka MM, Merski M, Bandyra KJ, Górna MW
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
96 |
nm3 |
|
|
|
|
|
|
|
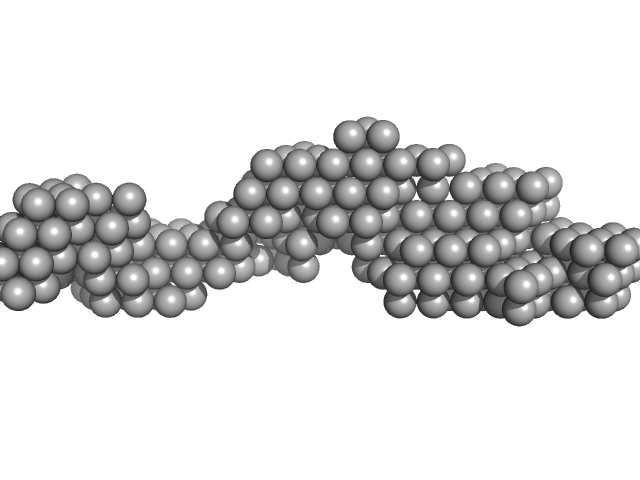
| Sample: |
Fas-activated serine/threonine kinase (amino acids 169-549) monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 5% (v/v) Glycerol, 200 mM Ammonium Sulphate, 50 mM NaCl, 0.5 mM TCEP, 10 mM Glutamic acid, 10 mM Arginine;, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 31
|
Human FASTK preferentially binds single-stranded and G-rich RNA.
FEBS J (2025)
Dawidziak DM, Dzadz DA, Kuska MI, Kanavalli M, Klimecka MM, Merski M, Bandyra KJ, Górna MW
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
BC120 RNA monomer monomer, 39 kDa RNA
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 22
|
Alu RNA pseudoknot alterations influence SRP9/SRP14 association.
RNA (2025)
Gussakovsky D, Brown MJF, Pereira HS, Meier M, Padilla-Meier GP, Black NA, Booy EP, Stetefeld J, Patel TR, McKenna SA
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
57 |
nm3 |
|
|