|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
20 mM PBS, 5 %(v/v) glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 5
|
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
J Biol Chem (2019)
Leth JM, Mertens HDT, Leth-Espensen KZ, Jørgensen TJD, Ploug M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
Urokinase-type plasminogen activator (Amino Terminal Fragment) monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
20 mM PBS, 5 %(v/v) glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 5
|
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
J Biol Chem (2019)
Leth JM, Mertens HDT, Leth-Espensen KZ, Jørgensen TJD, Ploug M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
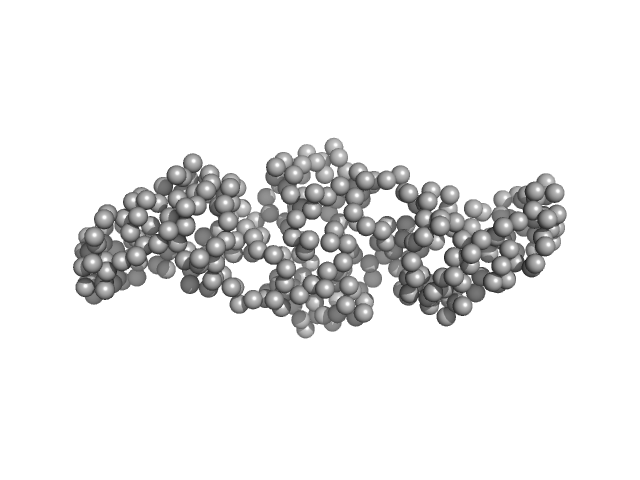
|
| Sample: |
Gliding motility protein MglB dimer, 34 kDa Myxococcus xanthus protein
|
| Buffer: |
150 mM NaCl, 1 mM DTT, 20 mM Tris-HCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Oct 9
|
MglA functions as a three-state GTPase to control movement reversals of Myxococcus xanthus.
Nat Commun 10(1):5300 (2019)
Galicia C, Lhospice S, Varela PF, Trapani S, Zhang W, Navaza J, Herrou J, Mignot T, Cherfils J
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
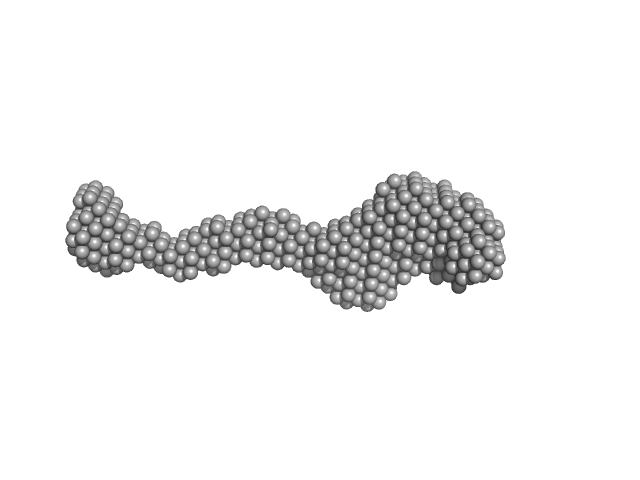
|
| Sample: |
Lysine-specific demethylase 5B monomer, 176 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 300 mM NaCl, 5% (v/v) glycerol, 1mM DTT, pH: 7.7 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with GeniX3D, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Oct 24
|
Molecular architecture of the Jumonji C family histone demethylase KDM5B.
Sci Rep 9(1):4019 (2019)
Dorosz J, Kristensen LH, Aduri NG, Mirza O, Lousen R, Bucciarelli S, Mehta V, Sellés-Baiget S, Solbak SMØ, Bach A, Mesa P, Hernandez PA, Montoya G, Nguyen TTTN, Rand KD, Boesen T, Gajhede M
|
| RgGuinier |
8.8 |
nm |
| Dmax |
26.9 |
nm |
|
|
|
|
|
|
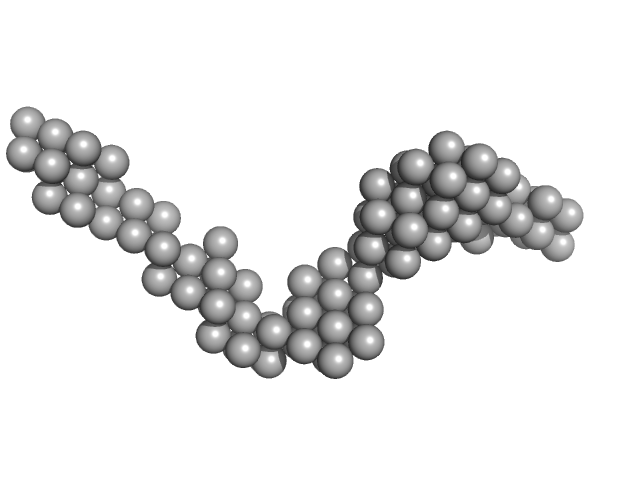
|
| Sample: |
Octo-repeat PrP mRNA dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Nov 4
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
7.4 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
|
|
|
|
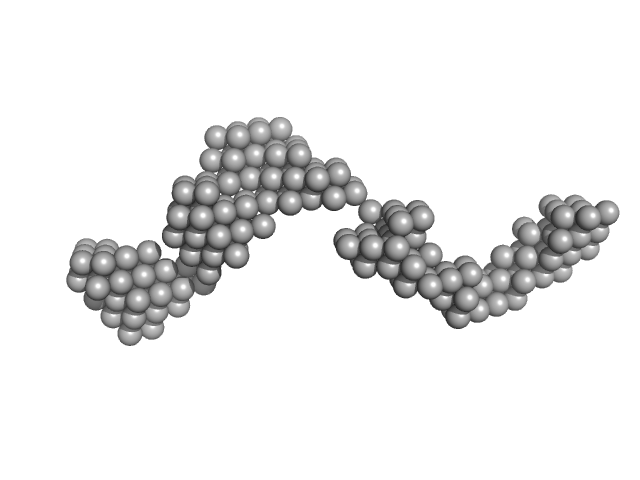
|
| Sample: |
Octo-repeat PrP mRNA dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
8.8 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
|
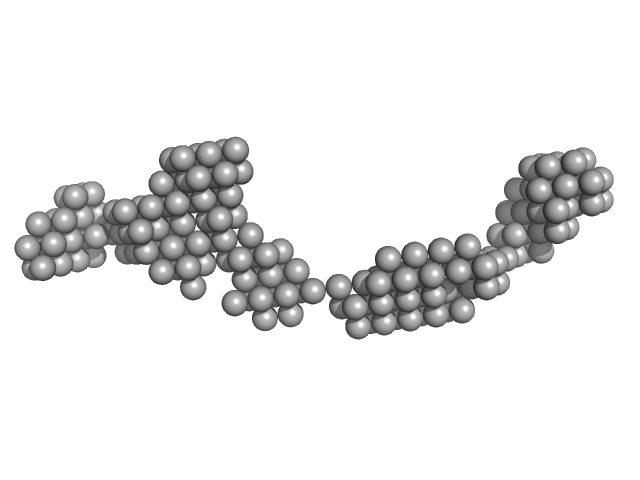
| Sample: |
Octo-repeat PrP mRNA dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM LiCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
9.0 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
|
|
|
|

|
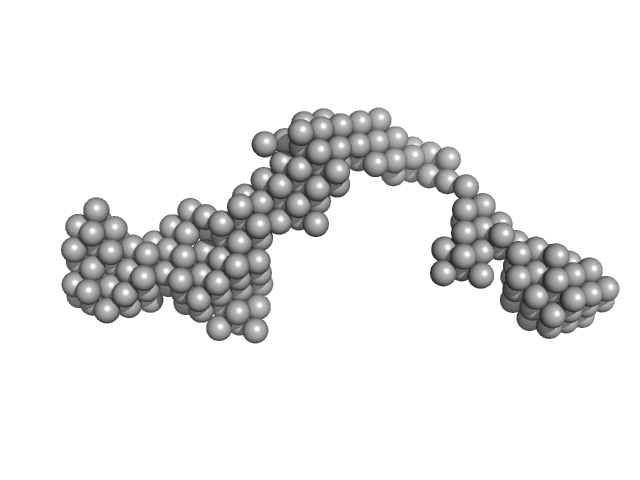
| Sample: |
Octo-repeat PrP mRNA dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM KCl and 1 mM PDS, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
9.1 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Octo-repeat PrP mRNA mutant dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
7.0 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Octo-repeat PrP mRNA mutant dimer, 144 kDa human PrP ORF RNA
|
| Buffer: |
10 mM Tris buffer with 100 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Jun 6
|
Octa-repeat domain of the mammalian prion protein mRNA forms stable A-helical hairpin structure rather than G-quadruplexes.
Sci Rep 9(1):2465 (2019)
Czech A, Konarev PV, Goebel I, Svergun DI, Wills PR, Ignatova Z
|
| RgGuinier |
9.0 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
210 |
nm3 |
|
|