|
|
|
|
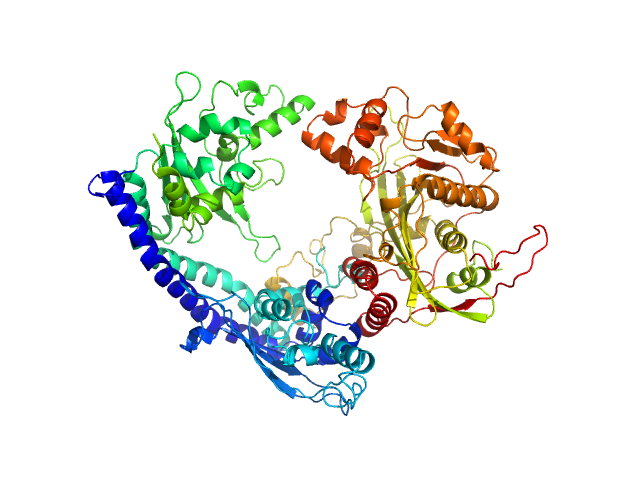
|
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
Histone H3 full-length monomer, 15 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 42% D2O, pH: 6.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 14
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity (Asf1-H3:H4-Rtt109-Vps75 protein complex, data for docking block selections)
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.5 |
nm |
|
|
|
|
|
|
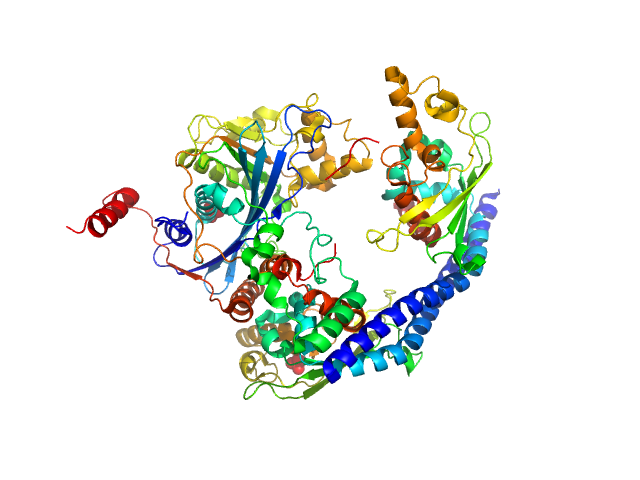
|
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
Histone H3 full-length monomer, 15 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 42% D2O, pH: 6.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 14
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity (Asf1-H3:H4-Rtt109-Vps75 protein complex, data for docking block selections)
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Proteasome activator PA28 heptamer, 232 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.5 mM TCEP, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Dec 2
|
The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis.
Nat Microbiol 4(11):1990-2000 (2019)
Xie SC, Metcalfe RD, Hanssen E, Yang T, Gillett DL, Leis AP, Morton CJ, Kuiper MJ, Parker MW, Spillman NJ, Wong W, Tsu C, Dick LR, Griffin MDW, Tilley L
|
| RgGuinier |
4.3 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
484 |
nm3 |
|
|
|
|
|
|
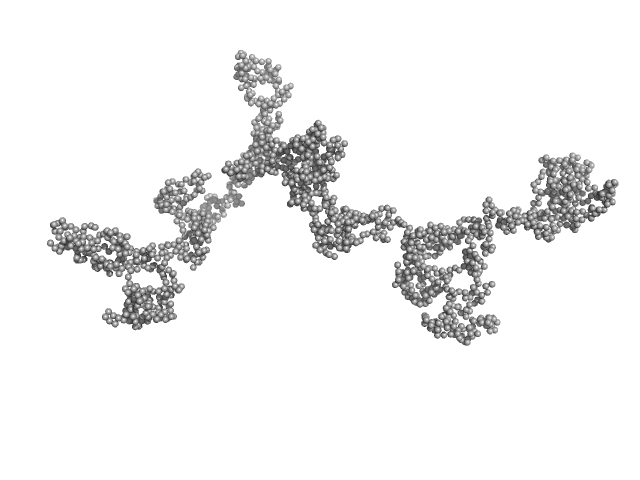
|
| Sample: |
Transcription intermediary factor 1-beta, TIF1b, KAP1, TRIM28 dimer, 183 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 6
|
KAP1 is an antiparallel dimer with a functional asymmetry.
Life Sci Alliance 2(4) (2019)
Fonti G, Marcaida MJ, Bryan LC, Träger S, Kalantzi AS, Helleboid PJ, Demurtas D, Tully MD, Grudinin S, Trono D, Fierz B, Dal Peraro M
|
| RgGuinier |
9.0 |
nm |
| Dmax |
38.0 |
nm |
| VolumePorod |
710 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Transcription intermediary factor 1-beta, TIF1b, KAP1, TRIM28, Fragment 23-418, RBCC domain dimer, 92 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 10 % Glycerol, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 6
|
KAP1 is an antiparallel dimer with a functional asymmetry.
Life Sci Alliance 2(4) (2019)
Fonti G, Marcaida MJ, Bryan LC, Träger S, Kalantzi AS, Helleboid PJ, Demurtas D, Tully MD, Grudinin S, Trono D, Fierz B, Dal Peraro M
|
| RgGuinier |
8.3 |
nm |
| Dmax |
37.0 |
nm |
| VolumePorod |
381 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Filamin A Ig-like domains 4-6 monomer, 32 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Feb 10
|
Critical Structural Defects Explain Filamin A Mutations Causing Mitral Valve Dysplasia.
Biophys J 117(8):1467-1475 (2019)
Haataja TJK, Capoulade R, Lecointe S, Hellman M, Merot J, Permi P, Pentikäinen U
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
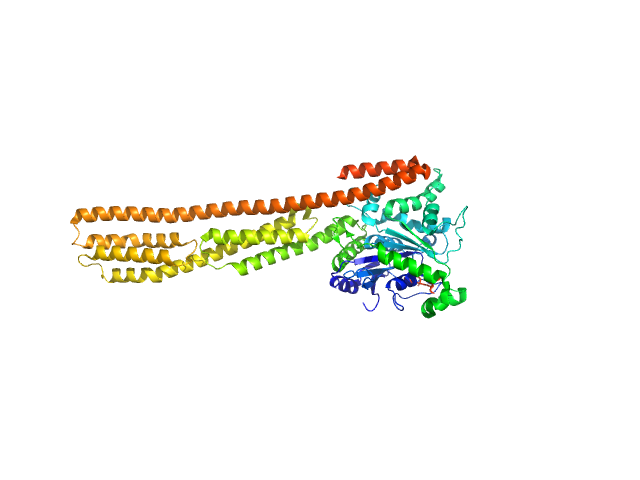
|
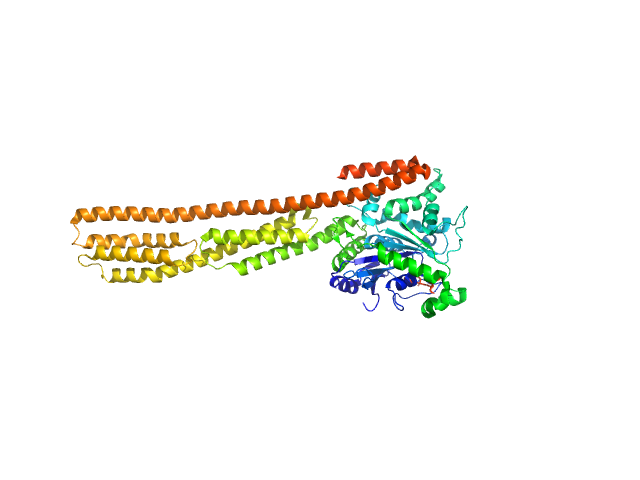
| Sample: |
Farnesylated human Guanylate-binding protein 1 monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 5 mM MgCl2, 150 mM NaCl, pH: 7.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 May 8
|
Farnesylation of human guanylate binding protein 1 as safety mechanism preventing structural rearrangements and uninduced dimerization.
FEBS J (2019)
Lorenz C, Ince S, Zhang T, Cousin A, Batra-Safferling R, Nagel-Steger L, Herrmann C, Stadler AM
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
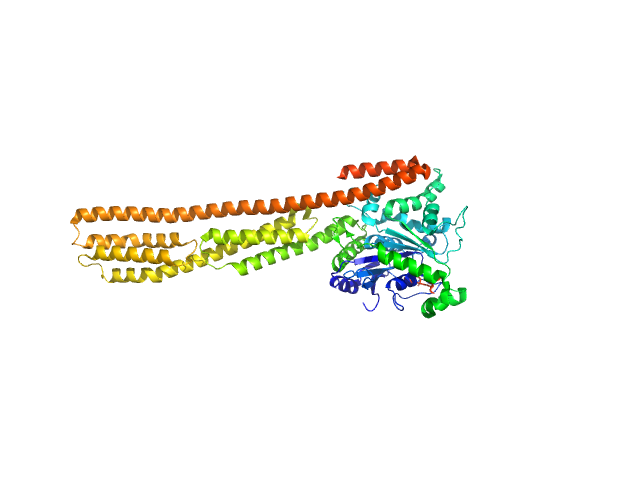
| Sample: |
Human Guanylate-binding protein 1 monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 5 mM MgCl2, 150 mM NaCl, pH: 7.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 May 8
|
Farnesylation of human guanylate binding protein 1 as safety mechanism preventing structural rearrangements and uninduced dimerization.
FEBS J (2019)
Lorenz C, Ince S, Zhang T, Cousin A, Batra-Safferling R, Nagel-Steger L, Herrmann C, Stadler AM
|
| RgGuinier |
5.1 |
nm |
| Dmax |
27.4 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
|
|
|
|
|
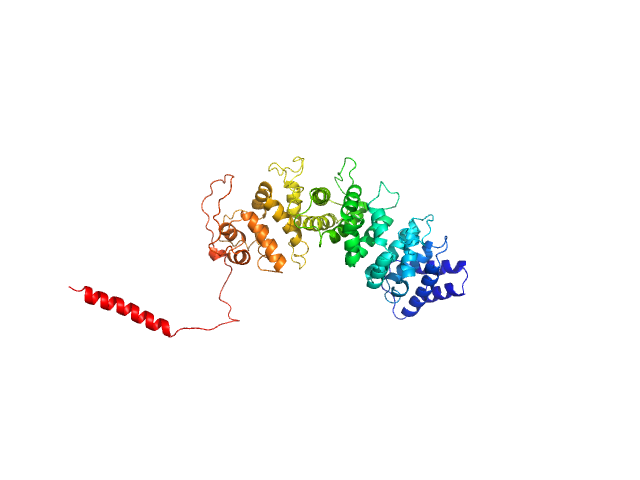
| Sample: |
Human Guanylate-binding protein 1 monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 5 mM MgCl2, 150 mM NaCl, 0.2 mM GppNHp, pH: 7.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 May 8
|
Farnesylation of human guanylate binding protein 1 as safety mechanism preventing structural rearrangements and uninduced dimerization.
FEBS J (2019)
Lorenz C, Ince S, Zhang T, Cousin A, Batra-Safferling R, Nagel-Steger L, Herrmann C, Stadler AM
|
| RgGuinier |
5.5 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Resistance to inhibitors of cholinesterase 8 homolog A monomer, 56 kDa Bos taurus protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 5 % glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Oct 27
|
Structural underpinnings of Ric8A function as a G-protein α-subunit chaperone and guanine-nucleotide exchange factor.
Nat Commun 10(1):3084 (2019)
Srivastava D, Gakhar L, Artemyev NO
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
88 |
nm3 |
|
|