|
|
|
|
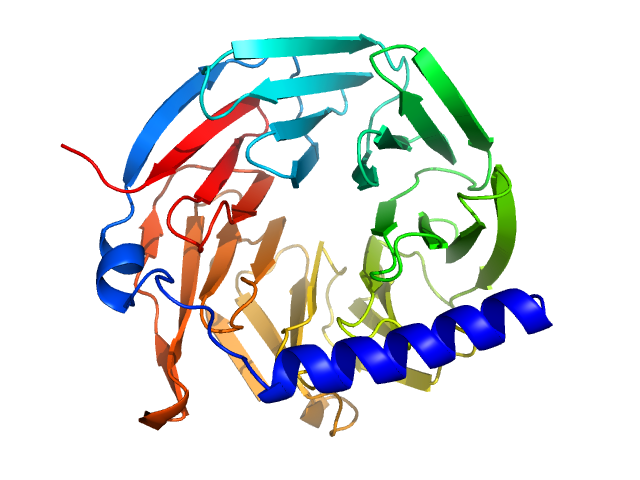
|
| Sample: |
Hypothetical protein CTHT_0072540 monomer, 35 kDa Chaetomium thermophilum protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 2 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jul 19
|
Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
Mol Cell 69(6):979-992.e6 (2018)
de Moura TR, Mozaffari-Jovin S, Szabó CZK, Schmitzová J, Dybkov O, Cretu C, Kachala M, Svergun D, Urlaub H, Lührmann R, Pena V
|
| RgGuinier |
2.3 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Deoxyuridine 5'-triphosphate nucleotidohydrolase trimer, 54 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 300 mM NaCl 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 9
|
Structural model of human dUTPase in complex with a novel proteinaceous inhibitor.
Sci Rep 8(1):4326 (2018)
Nyíri K, Mertens HDT, Tihanyi B, Nagy GN, Kőhegyi B, Matejka J, Harris MJ, Szabó JE, Papp-Kádár V, Németh-Pongrácz V, Ozohanics O, Vékey K, Svergun DI, Borysik AJ, Vértessy BG
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|
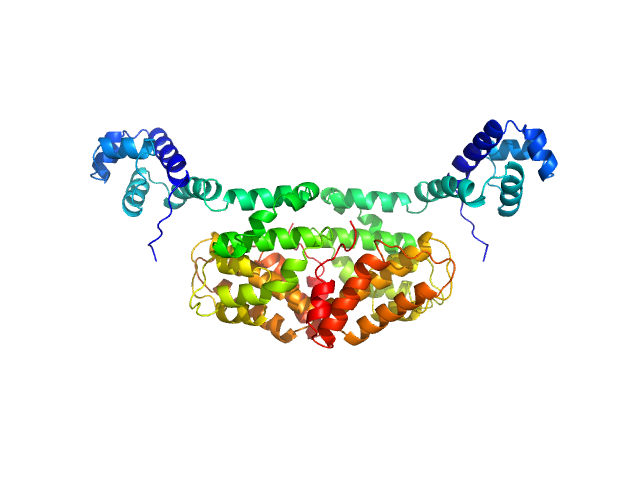
|
| Sample: |
SaPIbov1 pathogenicity island repressor dimer, 64 kDa Staphylococcus aureus protein
|
| Buffer: |
50 mM HEPES 300 mM NaCl 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 9
|
Structural model of human dUTPase in complex with a novel proteinaceous inhibitor.
Sci Rep 8(1):4326 (2018)
Nyíri K, Mertens HDT, Tihanyi B, Nagy GN, Kőhegyi B, Matejka J, Harris MJ, Szabó JE, Papp-Kádár V, Németh-Pongrácz V, Ozohanics O, Vékey K, Svergun DI, Borysik AJ, Vértessy BG
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
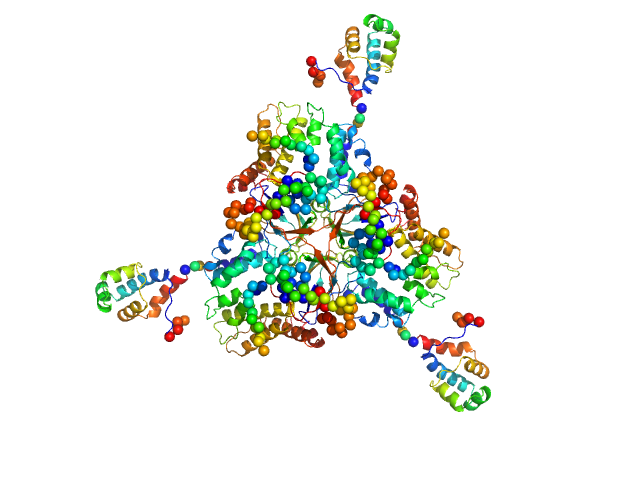
|
| Sample: |
Deoxyuridine 5'-triphosphate nucleotidohydrolase trimer, 54 kDa Homo sapiens protein
SaPIbov1 pathogenicity island repressor dimer, 64 kDa Staphylococcus aureus protein
|
| Buffer: |
50 mM HEPES 300 mM NaCl 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 10
|
Structural model of human dUTPase in complex with a novel proteinaceous inhibitor.
Sci Rep 8(1):4326 (2018)
Nyíri K, Mertens HDT, Tihanyi B, Nagy GN, Kőhegyi B, Matejka J, Harris MJ, Szabó JE, Papp-Kádár V, Németh-Pongrácz V, Ozohanics O, Vékey K, Svergun DI, Borysik AJ, Vértessy BG
|
| RgGuinier |
4.4 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
227 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Deoxyuridine 5'-triphosphate nucleotidohydrolase trimer, 54 kDa Homo sapiens protein
SaPIbov1 pathogenicity island repressor dimer, 64 kDa Staphylococcus aureus protein
|
| Buffer: |
50 mM HEPES 300 mM NaCl 5 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 10
|
Structural model of human dUTPase in complex with a novel proteinaceous inhibitor.
Sci Rep 8(1):4326 (2018)
Nyíri K, Mertens HDT, Tihanyi B, Nagy GN, Kőhegyi B, Matejka J, Harris MJ, Szabó JE, Papp-Kádár V, Németh-Pongrácz V, Ozohanics O, Vékey K, Svergun DI, Borysik AJ, Vértessy BG
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
|
|
|
|
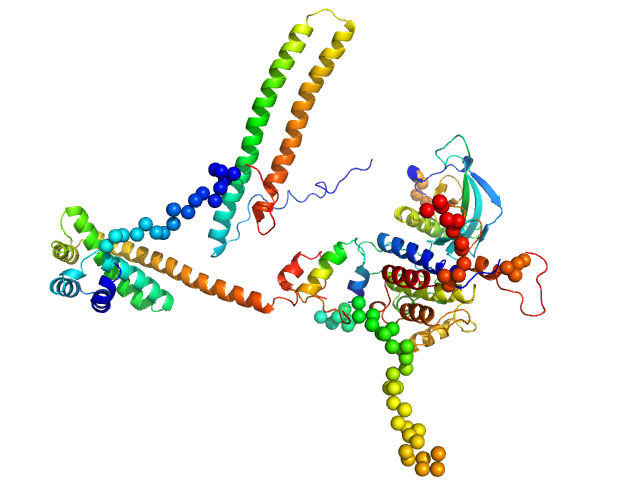
|
| Sample: |
Hsp90 co-chaperone Cdc37 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 5% (v/v) glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 10
|
Disease Variants of FGFR3 Reveal Molecular Basis for the Recognition and Additional Roles for Cdc37 in Hsp90 Chaperone System.
Structure 26(3):446-458.e8 (2018)
Bunney TD, Inglis AJ, Sanfelice D, Farrell B, Kerr CJ, Thompson GS, Masson GR, Thiyagarajan N, Svergun DI, Williams RL, Breeze AL, Katan M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|
|
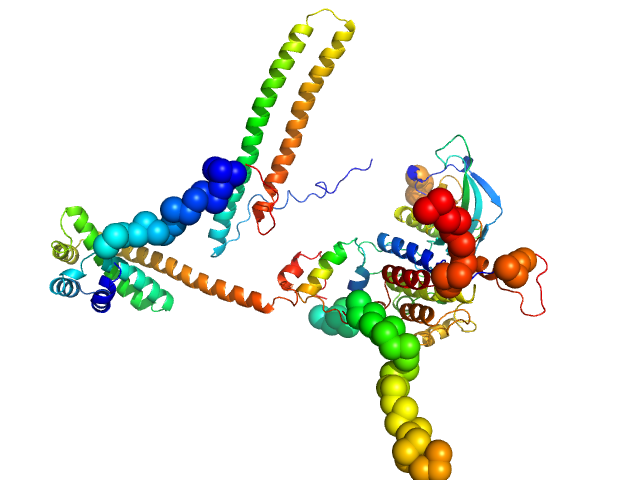
| Sample: |
Hsp90 co-chaperone Cdc37 monomer, 44 kDa Homo sapiens protein
Fibroblast growth factor receptor 3 monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 5% (v/v) glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 10
|
Disease Variants of FGFR3 Reveal Molecular Basis for the Recognition and Additional Roles for Cdc37 in Hsp90 Chaperone System.
Structure 26(3):446-458.e8 (2018)
Bunney TD, Inglis AJ, Sanfelice D, Farrell B, Kerr CJ, Thompson GS, Masson GR, Thiyagarajan N, Svergun DI, Williams RL, Breeze AL, Katan M
|
| RgGuinier |
4.7 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
161 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Fibroblast growth factor receptor 3 monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 5% (v/v) glycerol, 1 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 10
|
Disease Variants of FGFR3 Reveal Molecular Basis for the Recognition and Additional Roles for Cdc37 in Hsp90 Chaperone System.
Structure 26(3):446-458.e8 (2018)
Bunney TD, Inglis AJ, Sanfelice D, Farrell B, Kerr CJ, Thompson GS, Masson GR, Thiyagarajan N, Svergun DI, Williams RL, Breeze AL, Katan M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|

|
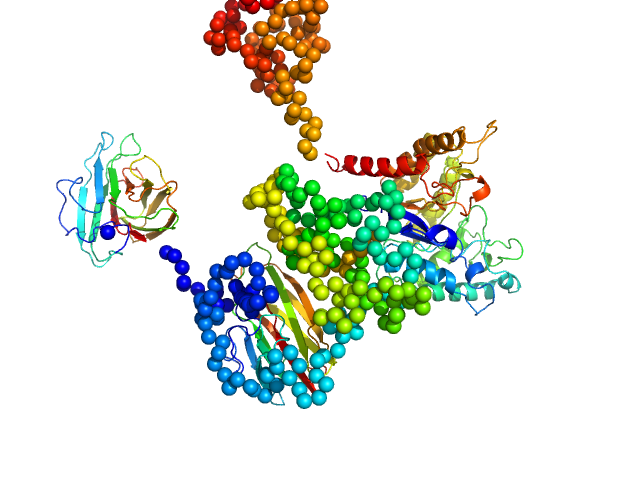
| Sample: |
Zinc finger and BTB domain-containing protein 38 monomer, 17 kDa Homo sapiens protein
Methylated C-terminal ZBTB38 binding sequence monomer, 17 kDa DNA
|
| Buffer: |
10 mM Tris, 1 mM tris(2-carboxy-ethyl)phosphine (TCEP), 0.05% NaN3, 10% D2O, pH: 6.8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, Department of Chemistry, University of Utah on 2016 Nov 11
|
The C-Terminal Zinc Fingers of ZBTB38 are Novel Selective Readers of DNA Methylation.
J Mol Biol 430(3):258-271 (2018)
Pozner A, Hudson NO, Trewhella J, Terooatea TW, Miller SA, Buck-Koehntop BA
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Prebiotic-producing GH10 xylanase (RmXyn10A, full length) monomer, 108 kDa Rhodothermus marinus protein
|
| Buffer: |
50 mM TRIS, 0.5 M NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Mar 12
|
Structural insights of RmXyn10A - A prebiotic-producing GH10 xylanase with a non-conserved aglycone binding region.
Biochim Biophys Acta Proteins Proteom 1866(2):292-306 (2018)
Aronsson A, Güler F, Petoukhov MV, Crennell SJ, Svergun DI, Linares-Pastén JA, Nordberg Karlsson E
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
170 |
nm3 |
|
|