|
|
|
|
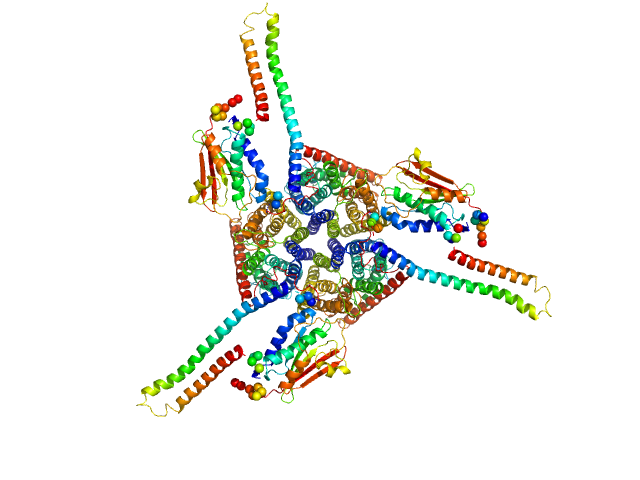
|
| Sample: |
Ammonium sensor/transducer trimer, 227 kDa Candidatus Kuenenia stuttgartiensis protein
|
| Buffer: |
20 mM Tris/HCl,100 mM NaCl, 5%(w/v) glycerol, 0.09%(w/v) CYMAL-5, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 1
|
Signaling ammonium across membranes through an ammonium sensor histidine kinase.
Nat Commun 9(1):164 (2018)
Pflüger T, Hernández CF, Lewe P, Frank F, Mertens H, Svergun D, Baumstark MW, Lunin VY, Jetten MSM, Andrade SLA
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
511 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cytohesin-3 monomer, 40 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 13
|
Structural Dynamics Control Allosteric Activation of Cytohesin Family Arf GTPase Exchange Factors.
Structure 26(1):106-117.e6 (2018)
Malaby AW, Das S, Chakravarthy S, Irving TC, Bilsel O, Lambright DG
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Nucleoplasmin core + A2 pentamer, 81 kDa Xenopus laevis protein
Histone H2A (ΔAla127) pentamer, 69 kDa Xenopus laevis protein
Histone H2B 1.1 (Ser33Thr) pentamer, 67 kDa Xenopus laevis protein
|
| Buffer: |
20 mM Tris. 150 mM NaCl, 1 mM EDTA, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2016 Jan 7
|
Dynamic intramolecular regulation of the histone chaperone nucleoplasmin controls histone binding and release.
Nat Commun 8(1):2215 (2017)
Warren C, Matsui T, Karp JM, Onikubo T, Cahill S, Brenowitz M, Cowburn D, Girvin M, Shechter D
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
402 |
nm3 |
|
|
|
|
|
|
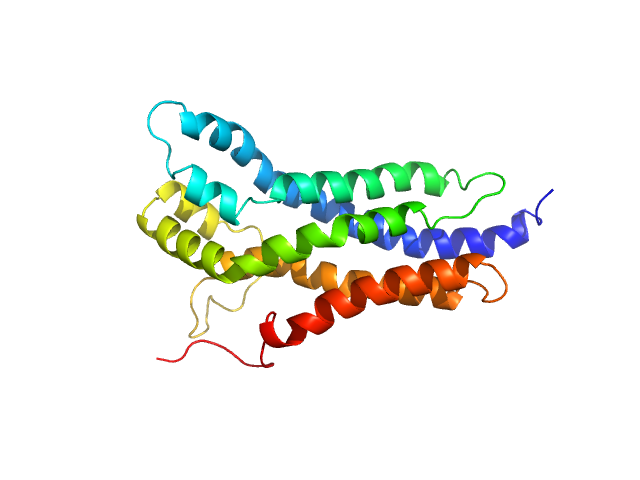
|
| Sample: |
BCR-ABL p210 fusion protein monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 5% Glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Oct 13
|
Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun 8(1):2101 (2017)
Reckel S, Gehin C, Tardivon D, Georgeon S, Kükenshöner T, Löhr F, Koide A, Buchner L, Panjkovich A, Reynaud A, Pinho S, Gerig B, Svergun D, Pojer F, Güntert P, Dötsch V, Koide S, Gavin AC, Hantschel O
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
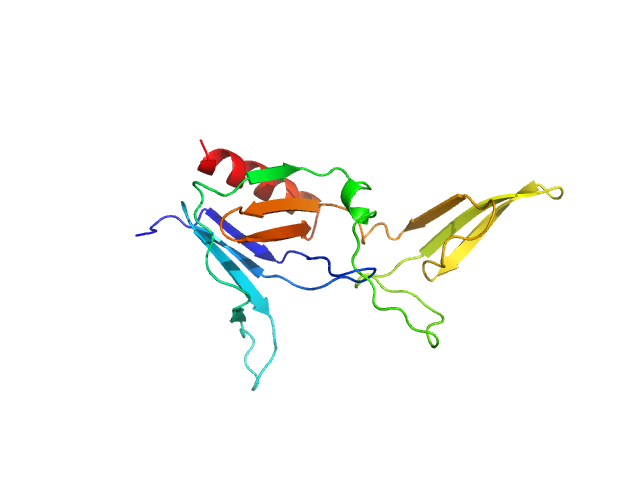
|
| Sample: |
BCR-ABL p210 fusion protein (PH domain) monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 5% Glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Nov 10
|
Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun 8(1):2101 (2017)
Reckel S, Gehin C, Tardivon D, Georgeon S, Kükenshöner T, Löhr F, Koide A, Buchner L, Panjkovich A, Reynaud A, Pinho S, Gerig B, Svergun D, Pojer F, Güntert P, Dötsch V, Koide S, Gavin AC, Hantschel O
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
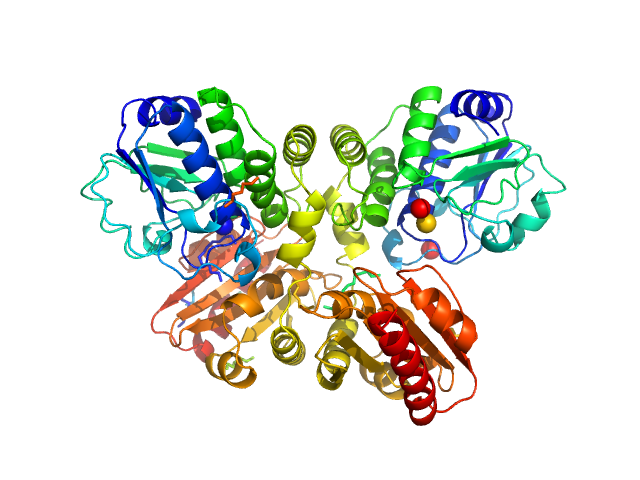
|
| Sample: |
Thermotoga maritima phosphodiesterase (wildtype, TmPDE, TM1595) dimer, 76 kDa Thermotoga maritima protein
|
| Buffer: |
20mM Tris, 200 mM NaCl, 5%(v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 23
|
Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure 25(12):1887-1897.e4 (2017)
Drexler DJ, Müller M, Rojas-Cordova CA, Bandera AM, Witte G
|
| RgGuinier |
2.7 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|
|
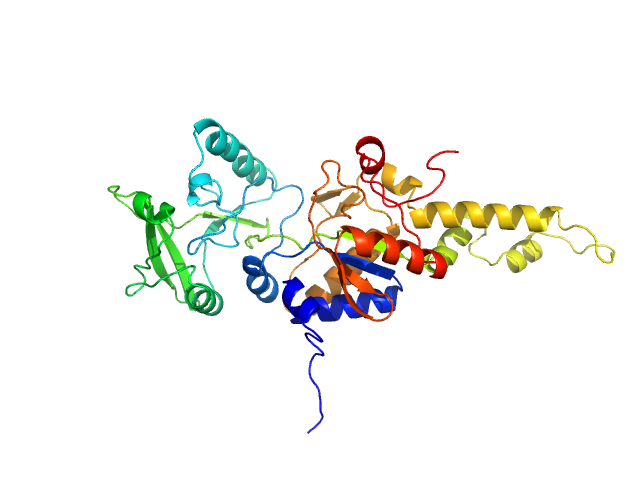
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 39 kDa Bordetella pertussis protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 4 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 19
|
Calmodulin fishing with a structurally disordered bait triggers CyaA catalysis.
PLoS Biol 15(12):e2004486 (2017)
O'Brien DP, Durand D, Voegele A, Hourdel V, Davi M, Chamot-Rooke J, Vachette P, Brier S, Ladant D, Chenal A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
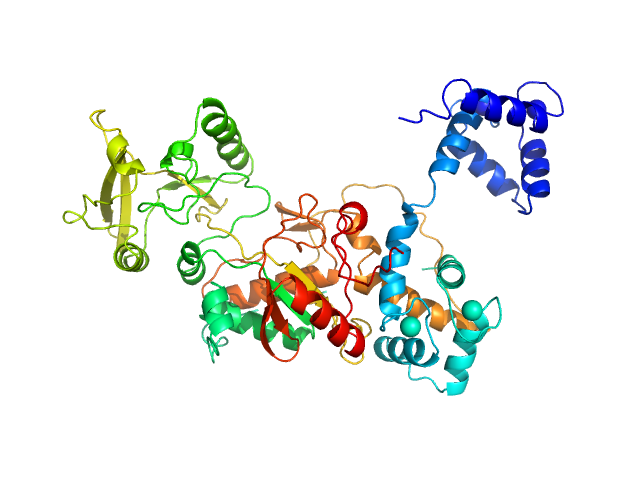
|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 39 kDa Bordetella pertussis protein
Calmodulin monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 4 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 19
|
Calmodulin fishing with a structurally disordered bait triggers CyaA catalysis.
PLoS Biol 15(12):e2004486 (2017)
O'Brien DP, Durand D, Voegele A, Hourdel V, Davi M, Chamot-Rooke J, Vachette P, Brier S, Ladant D, Chenal A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Sortilin, also: Neurotensin-receptor 3 dimer, 153 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES pH 7.4, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 17
|
Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun 8(1):1708 (2017)
Leloup N, Lössl P, Meijer DH, Brennich M, Heck AJR, Thies-Weesie DME, Janssen BJC
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
|
|
|
|
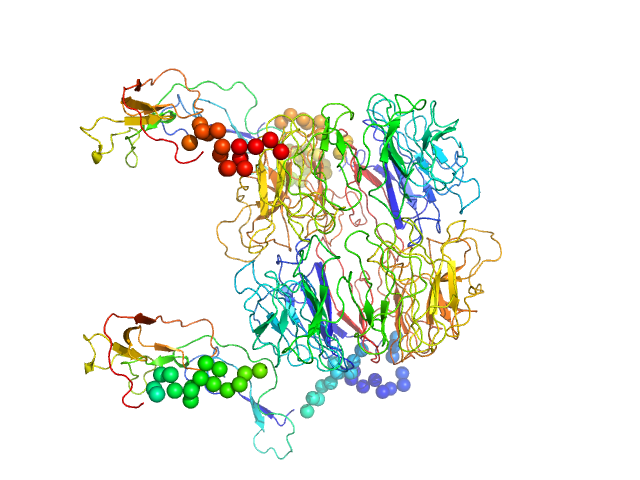
|
| Sample: |
Sortilin, also: Neurotensin-receptor 3 dimer, 153 kDa Mus musculus protein
|
| Buffer: |
25 mM MES pH 5.5, 150 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 17
|
Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun 8(1):1708 (2017)
Leloup N, Lössl P, Meijer DH, Brennich M, Heck AJR, Thies-Weesie DME, Janssen BJC
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
336 |
nm3 |
|
|