|
|
|
|
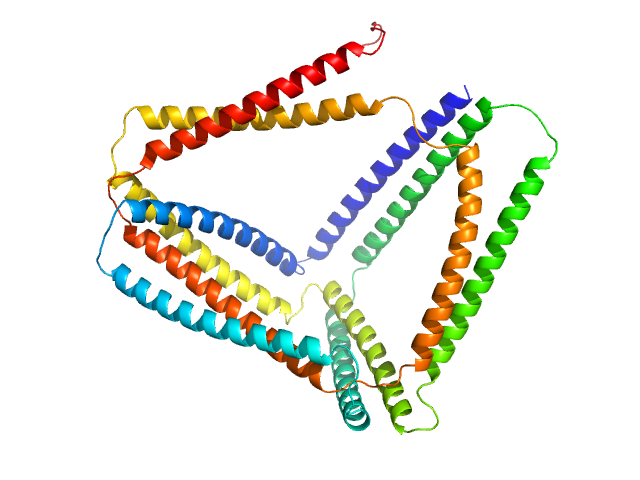
|
| Sample: |
TET12(1.10)S-c6 monomer, 55 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 7
|
Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo.
Nat Biotechnol 35(11):1094-1101 (2017)
Ljubetič A, Lapenta F, Gradišar H, Drobnak I, Aupič J, Strmšek Ž, Lainšček D, Hafner-Bratkovič I, Majerle A, Krivec N, Benčina M, Pisanski T, Veličković TĆ, Round A, Carazo JM, Melero R, Jerala R
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|
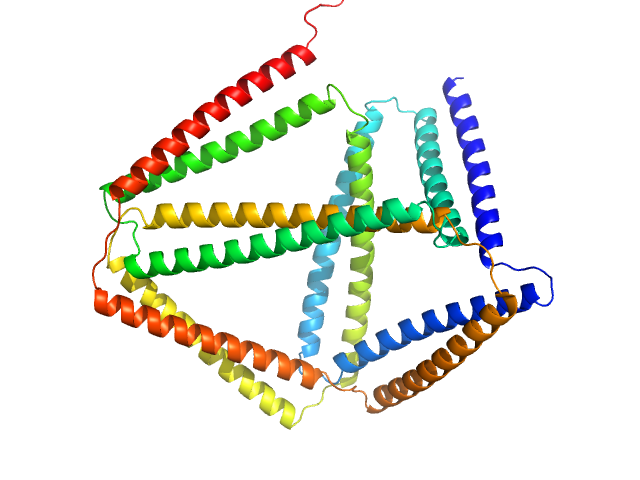
|
| Sample: |
TET12(1.6)S-c6b monomer, 54 kDa synthetic construct protein
|
| Buffer: |
20 mM Tris 150 mM NaCl 10% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Apr 7
|
Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo.
Nat Biotechnol 35(11):1094-1101 (2017)
Ljubetič A, Lapenta F, Gradišar H, Drobnak I, Aupič J, Strmšek Ž, Lainšček D, Hafner-Bratkovič I, Majerle A, Krivec N, Benčina M, Pisanski T, Veličković TĆ, Round A, Carazo JM, Melero R, Jerala R
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
114 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Rab family protein dimer, 130 kDa Chlorobaculum tepidum protein
|
| Buffer: |
20 mM HEPES 150 mM NaCl 5 mM MgCl2 5% Glycerol 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Feb 1
|
A homologue of the Parkinson's disease-associated protein LRRK2 undergoes a monomer-dimer transition during GTP turnover.
Nat Commun 8(1):1008 (2017)
Deyaert E, Wauters L, Guaitoli G, Konijnenberg A, Leemans M, Terheyden S, Petrovic A, Gallardo R, Nederveen-Schippers LM, Athanasopoulos PS, Pots H, Van Haastert PJM, Sobott F, Gloeckner CJ, Efremov R, Kortholt A, Versées W
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
211 |
nm3 |
|
|
|
|
|
|
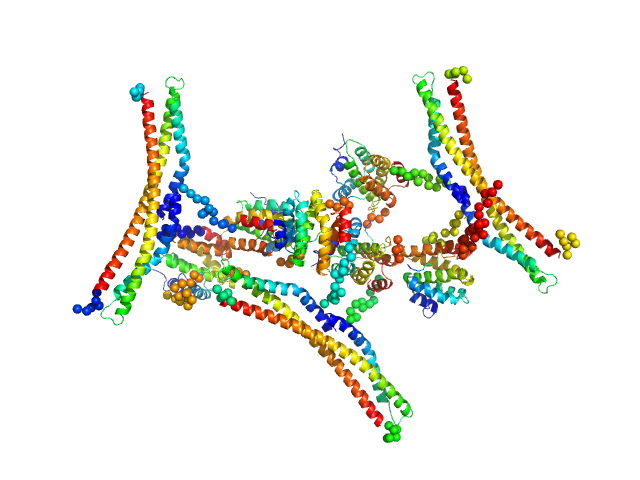
|
| Sample: |
Regulator of Ty1 transposition protein 103 dimer, 57 kDa Saccharomyces cerevisiae protein
DNA-directed RNA polymerase II subunit RPB1 monomer, 13 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
25 mM KH2PO4, 300 mM KCl, 10 mM BME, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2016 Apr 12
|
Structure and dynamics of the RNAPII CTDsome with Rtt103.
Proc Natl Acad Sci U S A 114(42):11133-11138 (2017)
Jasnovidova O, Klumpler T, Kubicek K, Kalynych S, Plevka P, Stefl R
|
| RgGuinier |
5.3 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
561 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
25 mM MOPS, 250 mM NaCl, 50 mM KCl, 2 mM TCEP, 0.1% NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Mar 9
|
2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update.
Acta Crystallogr D Struct Biol 73(Pt 9):710-728 (2017)
Trewhella J, Duff AP, Durand D, Gabel F, Guss JM, Hendrickson WA, Hura GL, Jacques DA, Kirby NM, Kwan AH, Pérez J, Pollack L, Ryan TM, Sali A, Schneidman-Duhovny D, Schwede T, Svergun DI, Sugiyama M, Tainer JA, Vachette P, Westbrook J, Whitten AE
|
| RgGuinier |
3.3 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
229 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Serum albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
25 mM MOPS, 250 mM NaCl, 50 mM KCl, 2 mM TCEP, 0.1% NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Mar 9
|
2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update.
Acta Crystallogr D Struct Biol 73(Pt 9):710-728 (2017)
Trewhella J, Duff AP, Durand D, Gabel F, Guss JM, Hendrickson WA, Hura GL, Jacques DA, Kirby NM, Kwan AH, Pérez J, Pollack L, Ryan TM, Sali A, Schneidman-Duhovny D, Schwede T, Svergun DI, Sugiyama M, Tainer JA, Vachette P, Westbrook J, Whitten AE
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
|
|
|
|
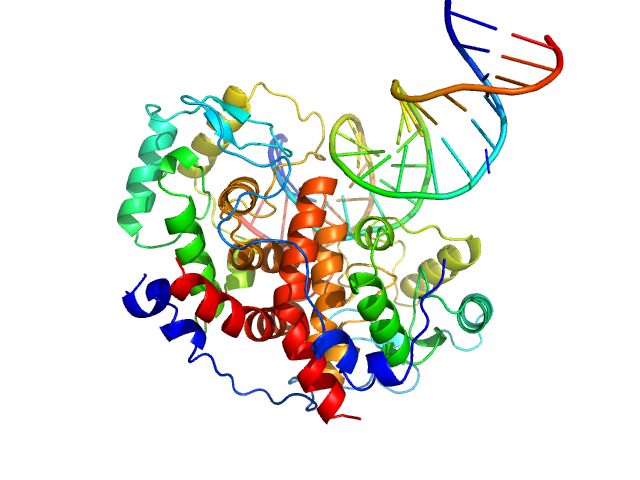
|
| Sample: |
Major viral transcription factor ICP4 dimer, 49 kDa Human alphaherpesvirus 1 protein
Immediate Early 3 dimer, 12 kDa DNA
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2015 Jul 27
|
The herpes viral transcription factor ICP4 forms a novel DNA recognition complex.
Nucleic Acids Res 45(13):8064-8078 (2017)
Tunnicliffe RB, Lockhart-Cairns MP, Levy C, Mould AP, Jowitt TA, Sito H, Baldock C, Sandri-Goldin RM, Golovanov AP
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
|
|
|
|
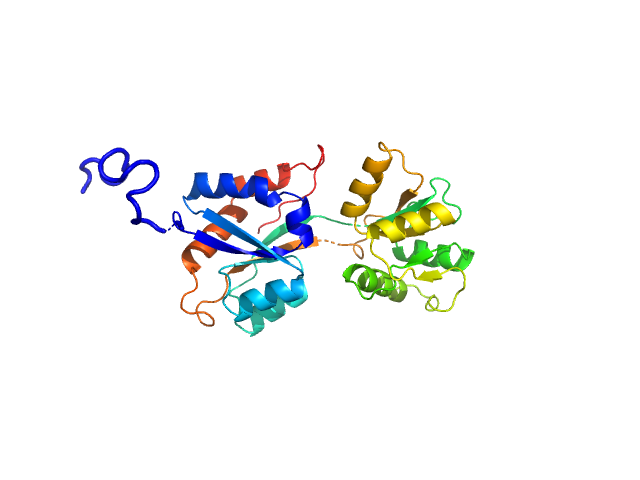
|
| Sample: |
ABC transporter periplasmic substrate-binding protein monomer, 30 kDa Desulfovibrio alaskensis protein
|
| Buffer: |
5 mM Tris-HCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 7
|
Highly selective tungstate transporter protein TupA from Desulfovibrio alaskensis G20.
Sci Rep 7(1):5798 (2017)
Otrelo-Cardoso AR, Nair RR, Correia MAS, Cordeiro RSC, Panjkovich A, Svergun DI, Santos-Silva T, Rivas MG
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
|
|
|
|
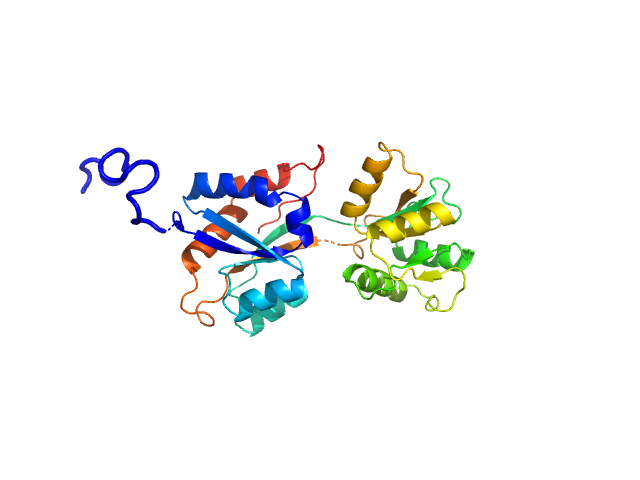
|
| Sample: |
ABC transporter periplasmic substrate-binding protein monomer, 30 kDa Desulfovibrio alaskensis protein
|
| Buffer: |
5 mM Tris-HCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Jun 21
|
Highly selective tungstate transporter protein TupA from Desulfovibrio alaskensis G20.
Sci Rep 7(1):5798 (2017)
Otrelo-Cardoso AR, Nair RR, Correia MAS, Cordeiro RSC, Panjkovich A, Svergun DI, Santos-Silva T, Rivas MG
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
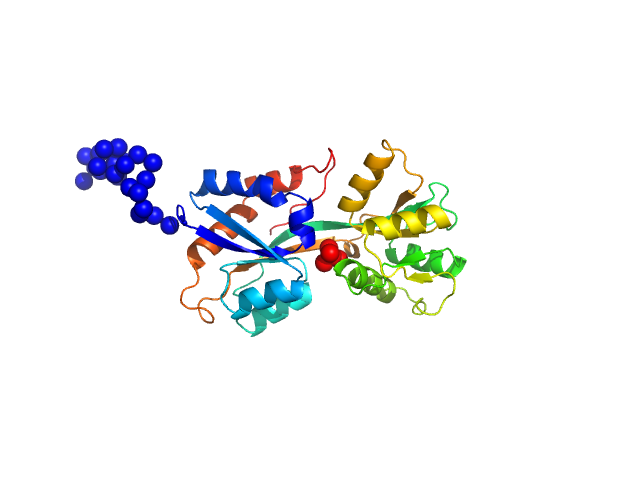
|
| Sample: |
ABC transporter periplasmic substrate-binding protein monomer, 30 kDa Desulfovibrio alaskensis protein
|
| Buffer: |
5 mM Tris-HCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 7
|
Highly selective tungstate transporter protein TupA from Desulfovibrio alaskensis G20.
Sci Rep 7(1):5798 (2017)
Otrelo-Cardoso AR, Nair RR, Correia MAS, Cordeiro RSC, Panjkovich A, Svergun DI, Santos-Silva T, Rivas MG
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|