|
|
|
|
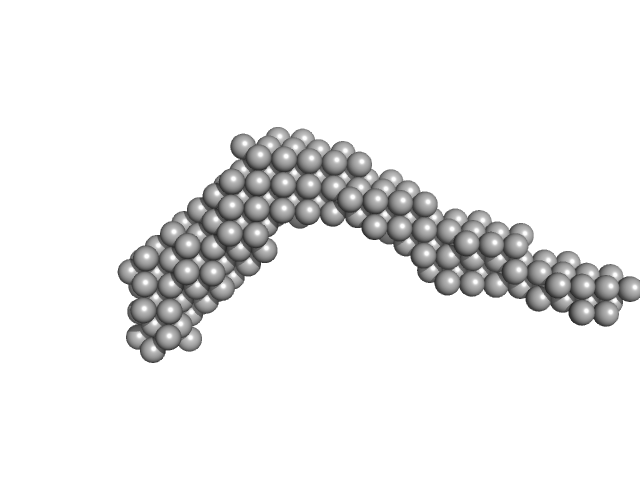
|
| Sample: |
AIR-3A tetramer, 26 kDa RNA
Interleukin-6 receptor subunit alpha dimer, 82 kDa Homo sapiens protein
|
| Buffer: |
water, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2014 Oct 1
|
Structure and target interaction of a G-quadruplex RNA-aptamer.
RNA Biol 13(10):973-987 (2016)
Szameit K, Berg K, Kruspe S, Valentini E, Magbanua E, Kwiatkowski M, Chauvot de Beauchêne I, Krichel B, Schamoni K, Uetrecht C, Svergun DI, Schlüter H, Zacharias M, Hahn U
|
| RgGuinier |
6.7 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
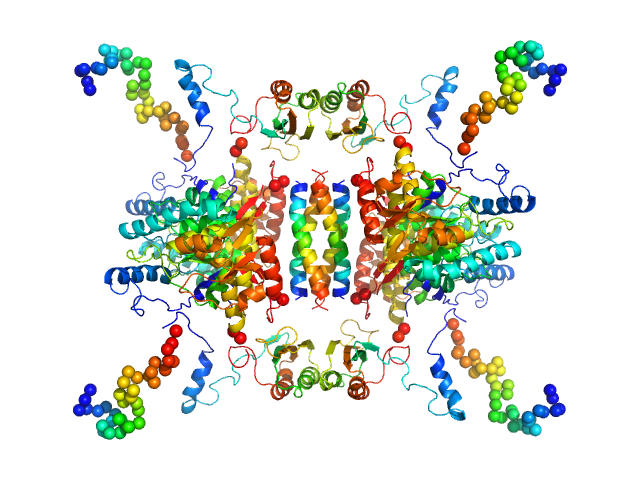
|
| Sample: |
Tyrosine hydroxylase, isoform 1 tetramer, 222 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES 200 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 14
|
Stable preparations of tyrosine hydroxylase provide the solution structure of the full-length enzyme.
Sci Rep 6:30390 (2016)
Bezem MT, Baumann A, Skjærven L, Meyer R, Kursula P, Martinez A, Flydal MI
|
| RgGuinier |
4.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
520 |
nm3 |
|
|
|
|
|
|
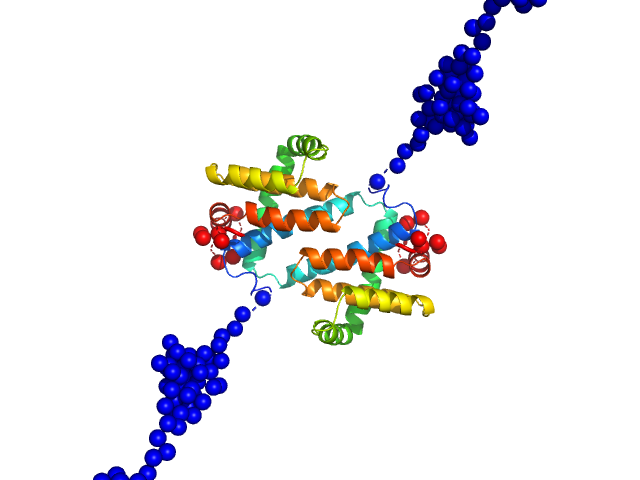
|
| Sample: |
MVA F1L antiapoptotic Bcl-2 viral protein dimer, 51 kDa Vaccinia virus protein
|
| Buffer: |
25 mM HEPES 150 mM NaCl 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Apr 15
|
The N Terminus of the Vaccinia Virus Protein F1L Is an Intrinsically Unstructured Region That Is Not Involved in Apoptosis Regulation.
J Biol Chem 291(28):14600-8 (2016)
Caria S, Marshall B, Burton RL, Campbell S, Pantaki-Eimany D, Hawkins CJ, Barry M, Kvansakul M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
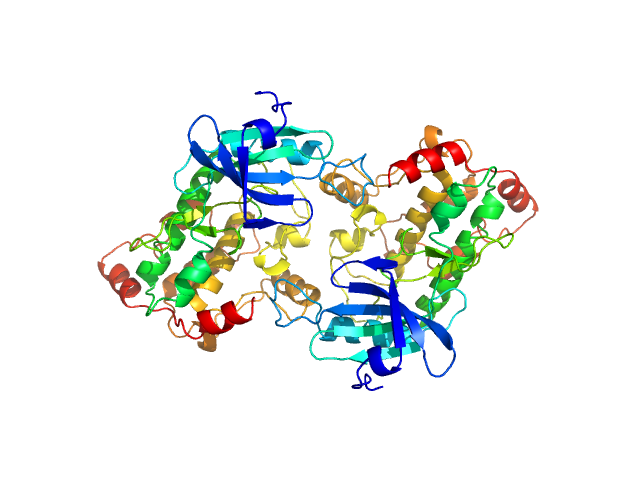
|
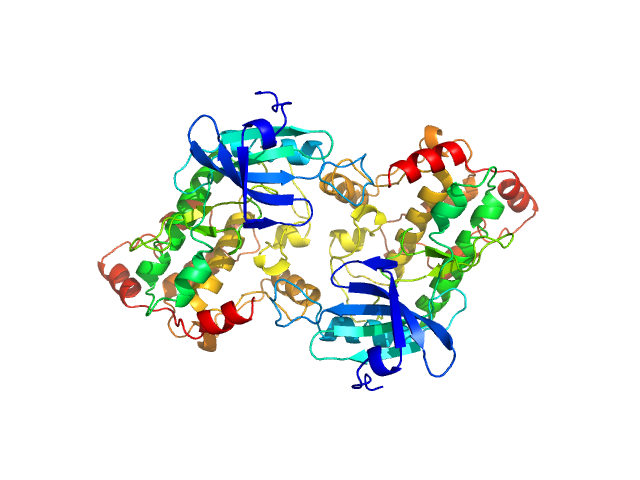
| Sample: |
Death associated protein kinase wild-type, 37 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 250 mM NaCl 5mM CaCl2 0.25 mM TCEP 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 18
|
Death-Associated Protein Kinase Activity Is Regulated by Coupled Calcium/Calmodulin Binding to Two Distinct Sites.
Structure 24(6):851-61 (2016)
Simon B, Huart AS, Temmerman K, Vahokoski J, Mertens HD, Komadina D, Hoffmann JE, Yumerefendi H, Svergun DI, Kursula P, Schultz C, McCarthy AA, Hart DJ, Wilmanns M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
|
|
|
|
|
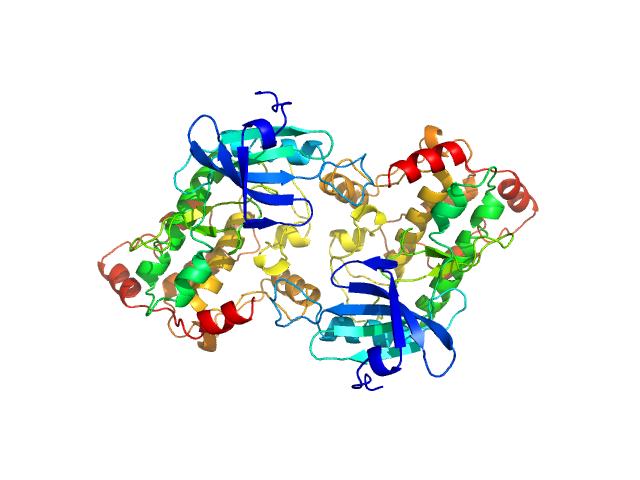
| Sample: |
Death associated protein kinase (D220K mutant), 37 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 250 mM NaCl 5mM CaCl2 0.25 mM TCEP 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 18
|
Death-Associated Protein Kinase Activity Is Regulated by Coupled Calcium/Calmodulin Binding to Two Distinct Sites.
Structure 24(6):851-61 (2016)
Simon B, Huart AS, Temmerman K, Vahokoski J, Mertens HD, Komadina D, Hoffmann JE, Yumerefendi H, Svergun DI, Kursula P, Schultz C, McCarthy AA, Hart DJ, Wilmanns M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
|
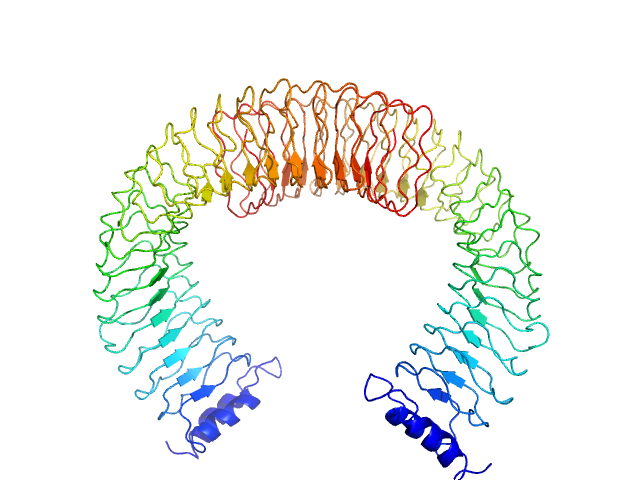
| Sample: |
Death associated protein kinase (Basic Loop mutant), 37 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 250 mM NaCl 5mM CaCl2 0.25 mM TCEP 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Dec 18
|
Death-Associated Protein Kinase Activity Is Regulated by Coupled Calcium/Calmodulin Binding to Two Distinct Sites.
Structure 24(6):851-61 (2016)
Simon B, Huart AS, Temmerman K, Vahokoski J, Mertens HD, Komadina D, Hoffmann JE, Yumerefendi H, Svergun DI, Kursula P, Schultz C, McCarthy AA, Hart DJ, Wilmanns M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
|
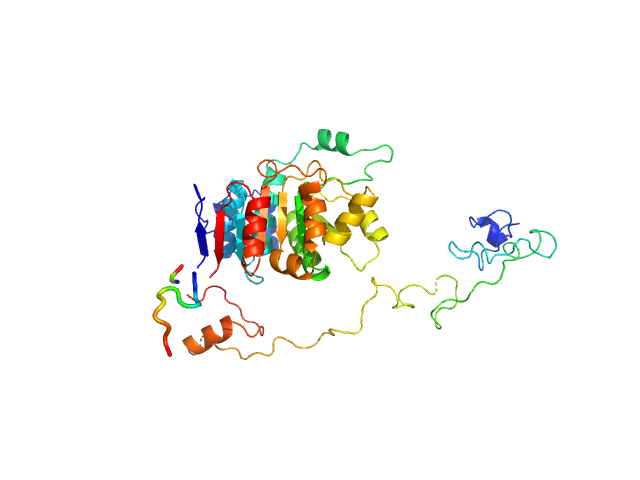
| Sample: |
Yersinia outer protein M (34-481) dimer, 101 kDa Yersinia enterocolitica protein
|
| Buffer: |
50 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 13
|
Immunosuppressive Yersinia Effector YopM Binds DEAD Box Helicase DDX3 to Control Ribosomal S6 Kinase in the Nucleus of Host Cells.
PLoS Pathog 12(6):e1005660 (2016)
Berneking L, Schnapp M, Rumm A, Trasak C, Ruckdeschel K, Alawi M, Grundhoff A, Kikhney AG, Koch-Nolte F, Buck F, Perbandt M, Betzel C, Svergun DI, Hentschke M, Aepfelbacher M
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
154 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DEAD box RNA helicase DDX3 (51-418) monomer, 42 kDa Yersinia enterocolitica protein
|
| Buffer: |
50 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 13
|
Immunosuppressive Yersinia Effector YopM Binds DEAD Box Helicase DDX3 to Control Ribosomal S6 Kinase in the Nucleus of Host Cells.
PLoS Pathog 12(6):e1005660 (2016)
Berneking L, Schnapp M, Rumm A, Trasak C, Ruckdeschel K, Alawi M, Grundhoff A, Kikhney AG, Koch-Nolte F, Buck F, Perbandt M, Betzel C, Svergun DI, Hentschke M, Aepfelbacher M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
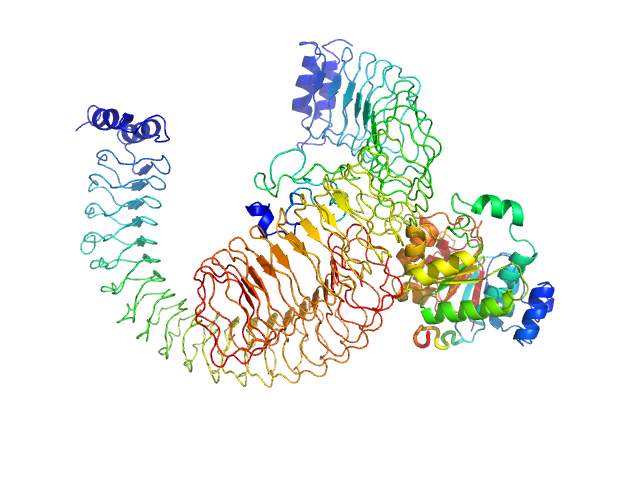
|
| Sample: |
Yersinia outer protein M (34-481) dimer, 101 kDa Yersinia enterocolitica protein
DEAD box RNA helicase DDX3 (51-418) monomer, 42 kDa Yersinia enterocolitica protein
|
| Buffer: |
50 mM HEPES 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 14
|
Immunosuppressive Yersinia Effector YopM Binds DEAD Box Helicase DDX3 to Control Ribosomal S6 Kinase in the Nucleus of Host Cells.
PLoS Pathog 12(6):e1005660 (2016)
Berneking L, Schnapp M, Rumm A, Trasak C, Ruckdeschel K, Alawi M, Grundhoff A, Kikhney AG, Koch-Nolte F, Buck F, Perbandt M, Betzel C, Svergun DI, Hentschke M, Aepfelbacher M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
E3 ubiquitin-protein ligase RNF8 dimer, 35 kDa Homo sapiens protein
Ubiquitin-conjugating enzyme E2 N double mutant (C87K, K92A) dimer, 36 kDa Homo sapiens protein
Polyubiquitin-C dimer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES 200 mM NaCl 0.01 mM ZnSO4 1 mM DTT, pH: 6.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 8
|
RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J Biol Chem 291(18):9396-410 (2016)
Hodge CD, Ismail IH, Edwards RA, Hura GL, Xiao AT, Tainer JA, Hendzel MJ, Glover JN
|
| RgGuinier |
4.5 |
nm |
| Dmax |
18.9 |
nm |
| VolumePorod |
119 |
nm3 |
|
|