|
|
|
|
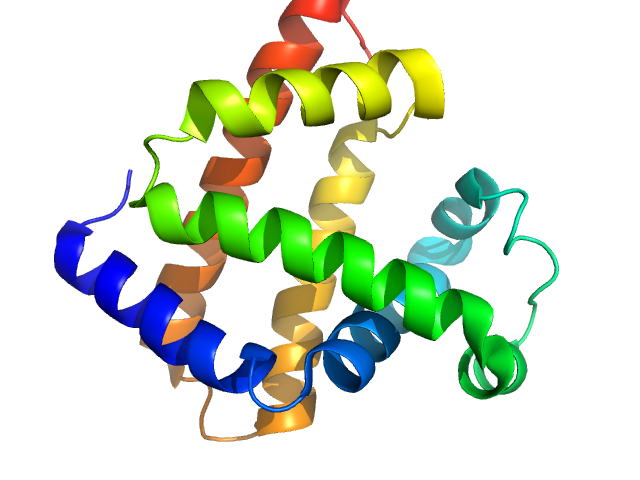
|
| Sample: |
Myoglobin monomer, 17 kDa Equus caballus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 20
|
Standard proteins
Darja Ruskule
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
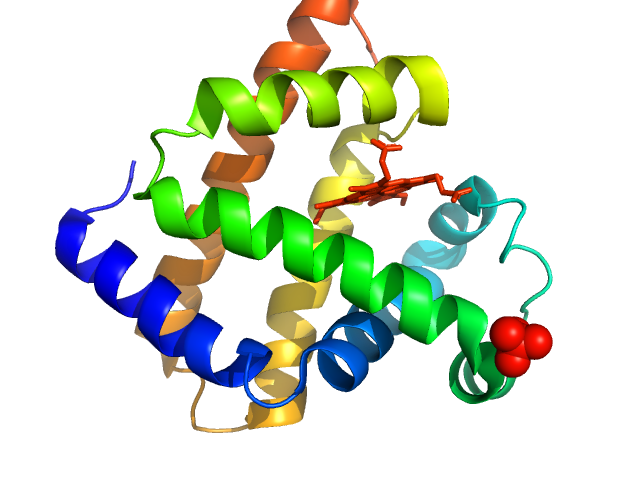
|
| Sample: |
Myoglobin monomer, 17 kDa Equus caballus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 20
|
Standard proteins
Darja Ruskule
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 18
|
Standard proteins
Darja Ruskule
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
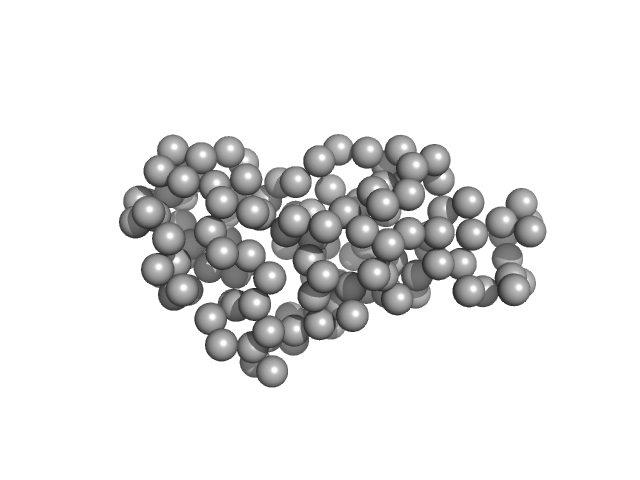
|
| Sample: |
Ribonuclease pancreatic monomer, 16 kDa Bos taurus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 18
|
Standard proteins
Darja Ruskule
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
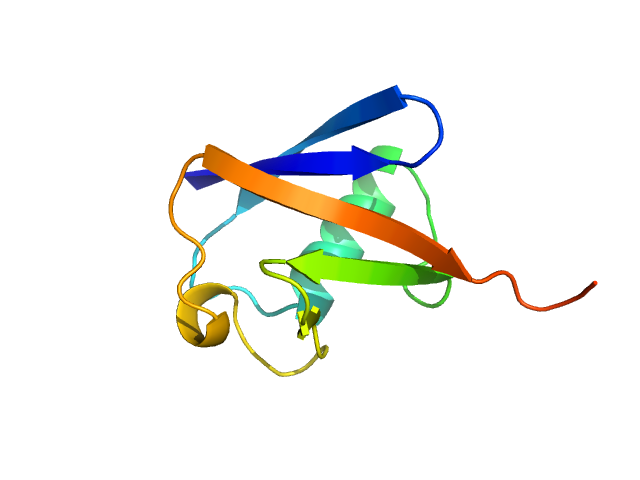
|
| Sample: |
Ubiquitin monomer, 9 kDa Bos taurus protein
|
| Buffer: |
40 mM Sodium acetate 150 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 18
|
Standard proteins
Darja Ruskule
|
| RgGuinier |
1.3 |
nm |
| Dmax |
4.9 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ribonuclease pancreatic monomer, 16 kDa Bos taurus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 18
|
Standard proteins
Darja Ruskule
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Xylose isomerase tetramer, 172 kDa Streptomyces rubiginosus protein
|
| Buffer: |
PBS, 50% Glycerol, 0.076 M NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Apr 24
|
Standard proteins
Erica Valentini
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
293 |
nm3 |
|
|
|
|
|
|
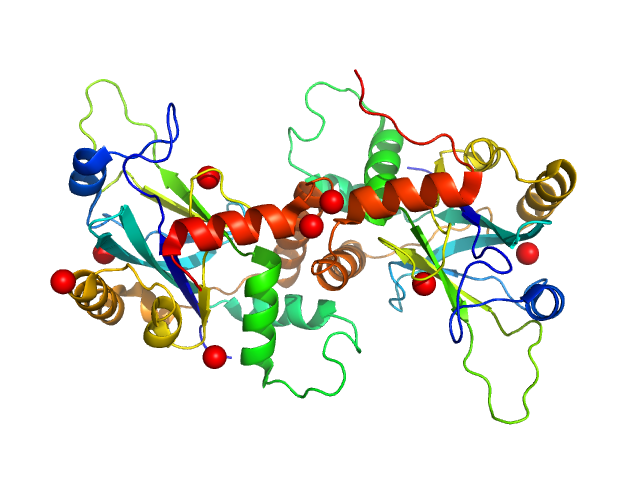
|
| Sample: |
VP24 dimer, 53 kDa Suid herpesvirus 1 protein
|
| Buffer: |
50 mM Tris/HCl 0.5 M NaCl 0.25 M imidazole 5% glycerol 50 mM urea 0.2 M MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 23
|
Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin pUL26N, a Herpesvirus Serine Protease.
PLoS Pathog 11(7):e1005045 (2015)
Zühlsdorf M, Werten S, Klupp BG, Palm GJ, Mettenleiter TC, Hinrichs W
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
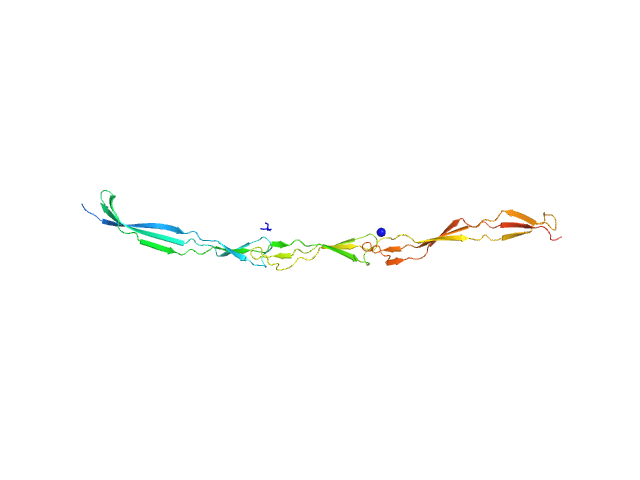
|
| Sample: |
Surface protein G monomer, 24 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
4.7 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
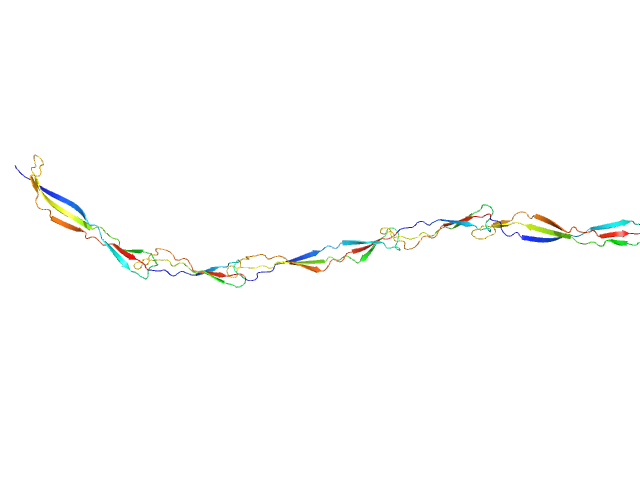
|
| Sample: |
Surface protein G monomer, 39 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
7.7 |
nm |
| Dmax |
30.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|