|
|
|
|
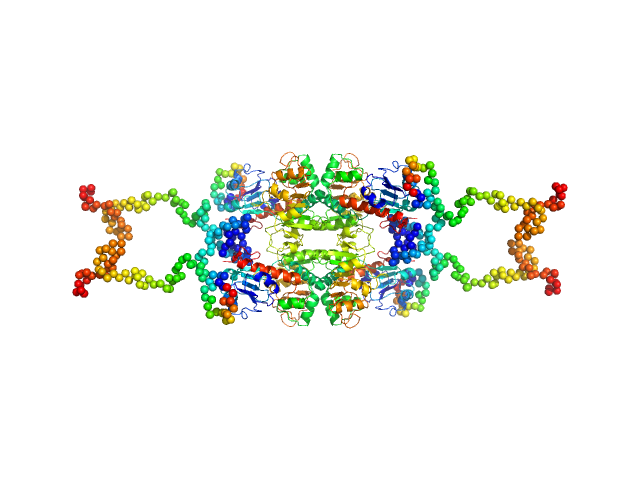
|
| Sample: |
C-terminal-binding protein 1 tetramer, 191 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 13
|
Master corepressor inactivation through multivalent SLiM-induced polymerization mediated by the oncogene suppressor RAI2.
Nat Commun 15(1):5241 (2024)
Goradia N, Werner S, Mullapudi E, Greimeier S, Bergmann L, Lang A, Mertens H, Węglarz A, Sander S, Chojnowski G, Wikman H, Ohlenschläger O, von Amsberg G, Pantel K, Wilmanns M
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
279 |
nm3 |
|
|
|
|
|
|
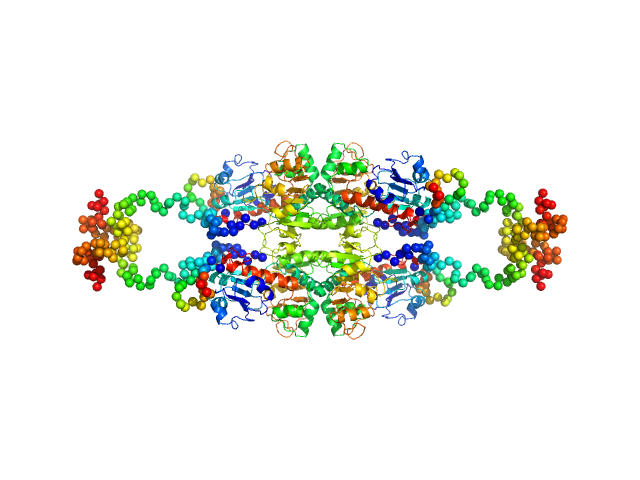
|
| Sample: |
C-terminal-binding protein 1 (C134Y, N138R, R141E, L150W), 48 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 13
|
Master corepressor inactivation through multivalent SLiM-induced polymerization mediated by the oncogene suppressor RAI2.
Nat Commun 15(1):5241 (2024)
Goradia N, Werner S, Mullapudi E, Greimeier S, Bergmann L, Lang A, Mertens H, Węglarz A, Sander S, Chojnowski G, Wikman H, Ohlenschläger O, von Amsberg G, Pantel K, Wilmanns M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
154 |
nm3 |
|
|
|
|
|
|
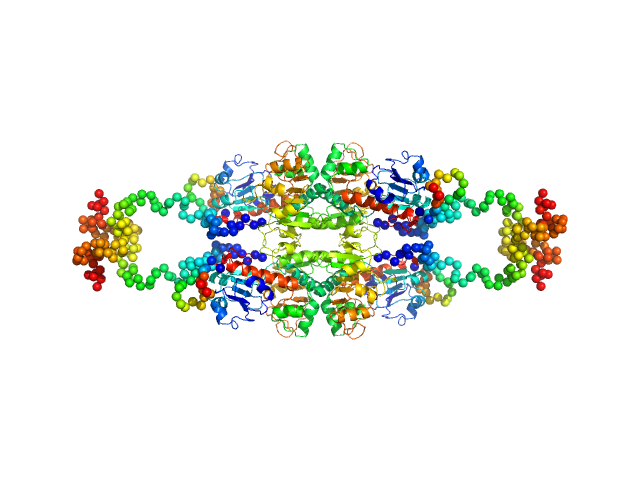
|
| Sample: |
C-terminal-binding protein 1 (R266A, D290A, E295A, H315A), 47 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 13
|
Master corepressor inactivation through multivalent SLiM-induced polymerization mediated by the oncogene suppressor RAI2.
Nat Commun 15(1):5241 (2024)
Goradia N, Werner S, Mullapudi E, Greimeier S, Bergmann L, Lang A, Mertens H, Węglarz A, Sander S, Chojnowski G, Wikman H, Ohlenschläger O, von Amsberg G, Pantel K, Wilmanns M
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
161 |
nm3 |
|
|
|
|
|
|
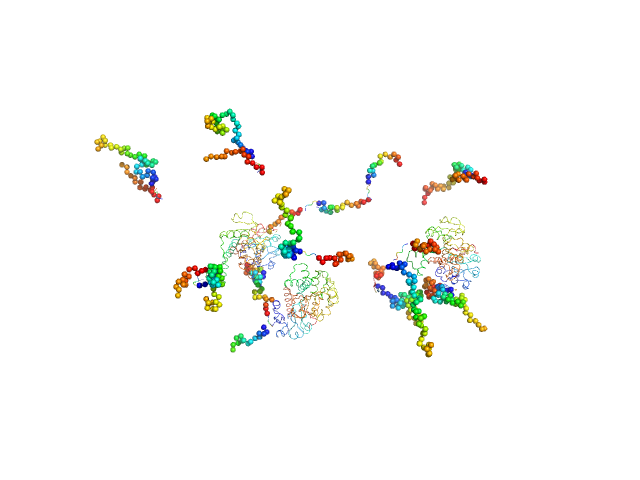
|
| Sample: |
C-terminal-binding protein 1 tetramer, 191 kDa Homo sapiens protein
Retinoic acid-induced protein 2 (303-465) monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 13
|
Master corepressor inactivation through multivalent SLiM-induced polymerization mediated by the oncogene suppressor RAI2.
Nat Commun 15(1):5241 (2024)
Goradia N, Werner S, Mullapudi E, Greimeier S, Bergmann L, Lang A, Mertens H, Węglarz A, Sander S, Chojnowski G, Wikman H, Ohlenschläger O, von Amsberg G, Pantel K, Wilmanns M
|
| RgGuinier |
7.4 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
978 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Alarmin release inhibitor (Δ1-62) monomer, 26 kDa Heligmosomoides polygyrus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, 5% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Feb 9
|
Structural basis for IL-33 recognition and its antagonism by the helminth effector protein HpARI2.
Nat Commun 15(1):5226 (2024)
Jamwal A, Colomb F, McSorley HJ, Higgins MK
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Alarmin release inhibitor (Δ1-125; N175Q, N190Q) monomer, 19 kDa Heligmosomoides polygyrus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, 5% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Feb 9
|
Structural basis for IL-33 recognition and its antagonism by the helminth effector protein HpARI2.
Nat Commun 15(1):5226 (2024)
Jamwal A, Colomb F, McSorley HJ, Higgins MK
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Alarmin release inhibitor (Δ1-62) monomer, 26 kDa Heligmosomoides polygyrus protein
Interleukin-33 (L179V) monomer, 18 kDa Mus musculus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, 5% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Feb 9
|
Structural basis for IL-33 recognition and its antagonism by the helminth effector protein HpARI2.
Nat Commun 15(1):5226 (2024)
Jamwal A, Colomb F, McSorley HJ, Higgins MK
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
|
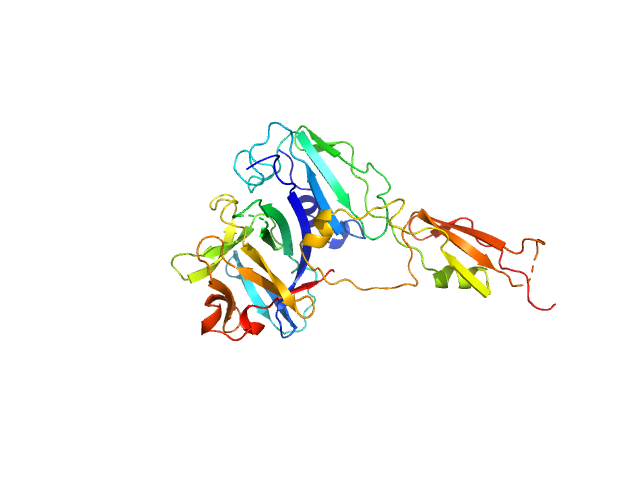
| Sample: |
Interleukin-33 (L179V) monomer, 18 kDa Mus musculus protein
Alarmin release inhibitor (Δ1-125; N175Q, N190Q) monomer, 19 kDa Heligmosomoides polygyrus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, 5% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Feb 9
|
Structural basis for IL-33 recognition and its antagonism by the helminth effector protein HpARI2.
Nat Commun 15(1):5226 (2024)
Jamwal A, Colomb F, McSorley HJ, Higgins MK
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
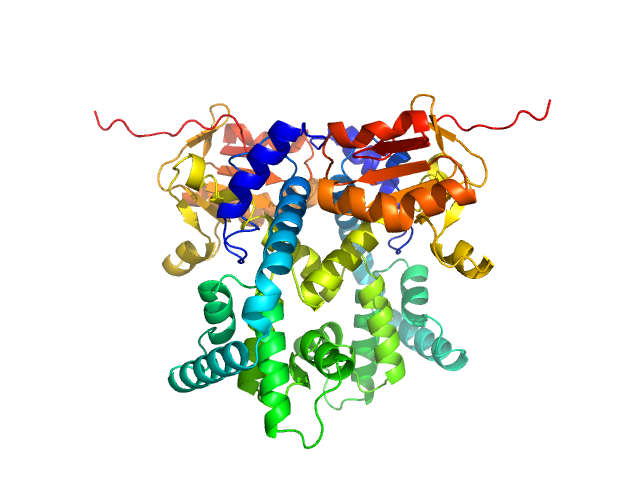
| Sample: |
CDAN1-interacting nuclease 1 dimer, 67 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2021 Sep 2
|
Structural insights into inherited anemia CDA-I: disease-associated mutations disrupt Codanin1-CDIN1 complex
Tomas Brom
|
| RgGuinier |
3.0 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
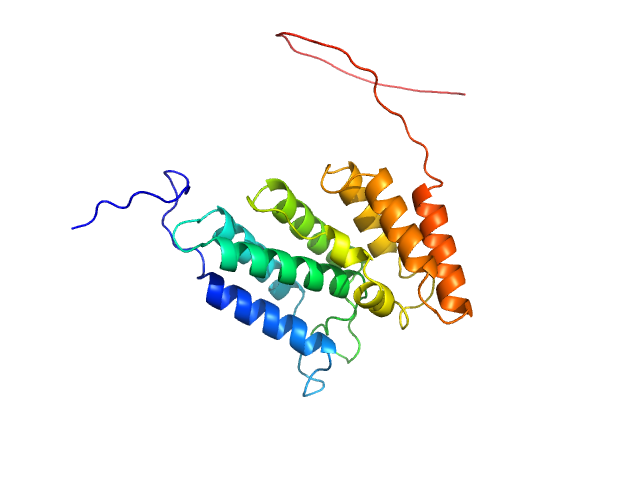
| Sample: |
Codanin-1 monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Mar 5
|
Structural insights into inherited anemia CDA-I: disease-associated mutations disrupt Codanin1-CDIN1 complex
Tomas Brom
|
| RgGuinier |
2.7 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
54 |
nm3 |
|
|