|
|
|
|
|
| Sample: |
Gramicidin S synthase 1 (W239S) monomer, 126 kDa Brevibacillus brevis protein
|
| Buffer: |
20 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Mar 14
|
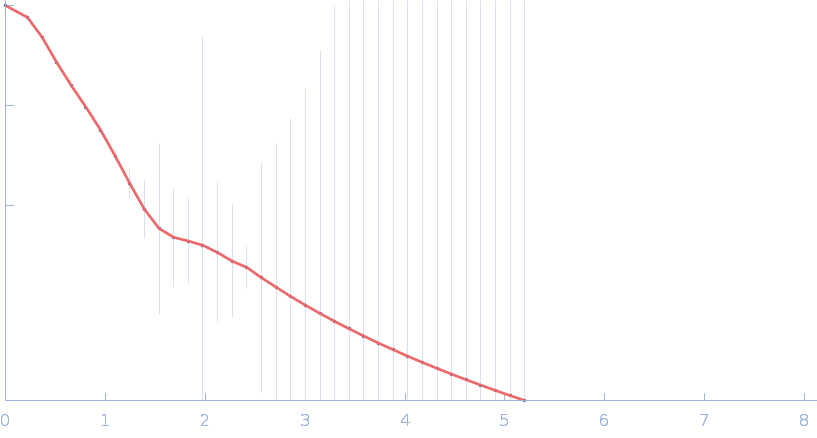
Resting state of GrsA is a combination of two constitutively inactive isoforms that manifest long-range allosteric effects in presence of aminoacid and ATP
Raktim Roy
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein A-I monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Mar 20
|
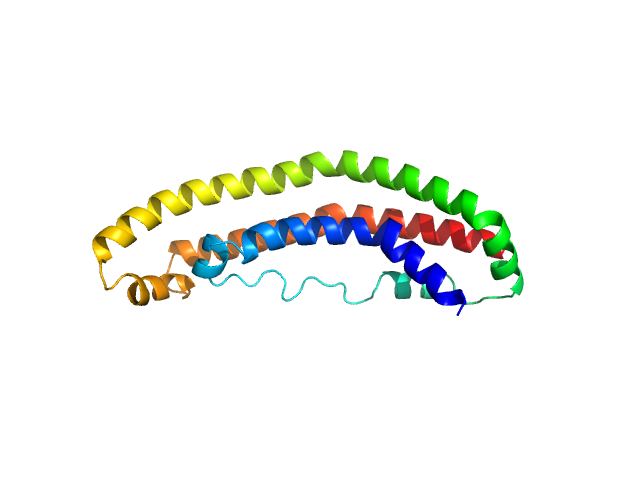
The structure of the apolipoprotein A-I monomer provides insights into its oligomerisation and lipid-binding mechanisms
Journal of Molecular Biology :169394 (2025)
Tou H, Rosenes Z, Khandokar Y, Zlatic C, Metcalfe R, Mok Y, Morton C, Gooley P, Griffin M
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Apolipoprotein A-I monomer, 28 kDa Homo sapiens protein
Antigen-binding fragment 55201 monomer, 45 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2021 Jun 19
|
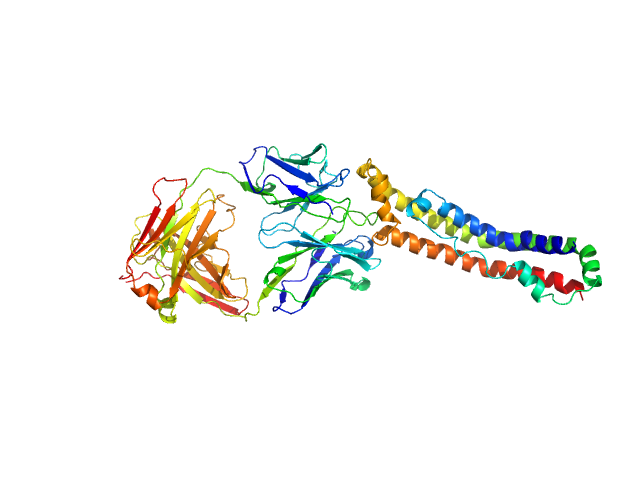
The structure of the apolipoprotein A-I monomer provides insights into its oligomerisation and lipid-binding mechanisms
Journal of Molecular Biology :169394 (2025)
Tou H, Rosenes Z, Khandokar Y, Zlatic C, Metcalfe R, Mok Y, Morton C, Gooley P, Griffin M
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
|
|
|
|
|
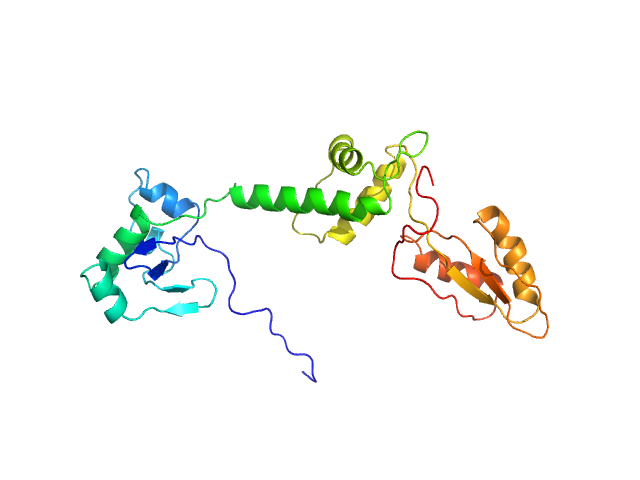
| Sample: |
Ribosome maturation protein SBDS monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 6
|
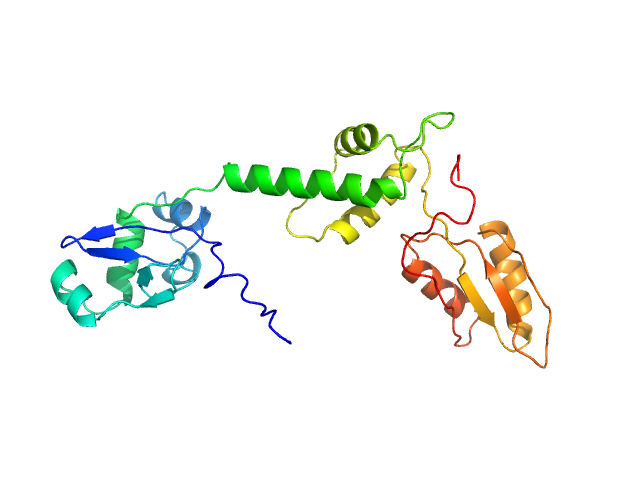
Structural Implications of Missense Point Mutations in Shwachman–Bodian–Diamond Syndrome Protein (SBDS): A Combined SAXS/MD Investigation
ACS Omega (2025)
Mattiotti G, Nanna V, Giulini M, Alberga D, Mangiatordi G, Sánchez-Puig N, Saviano M, Tubiana L, Potestio R, Lattanzi G, Siliqi D
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
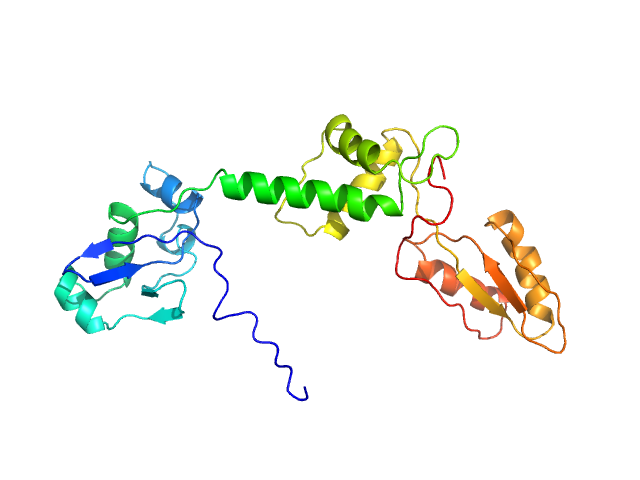
| Sample: |
Ribosome maturation protein SBDS (R19Q) monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 6
|
Structural Implications of Missense Point Mutations in Shwachman–Bodian–Diamond Syndrome Protein (SBDS): A Combined SAXS/MD Investigation
ACS Omega (2025)
Mattiotti G, Nanna V, Giulini M, Alberga D, Mangiatordi G, Sánchez-Puig N, Saviano M, Tubiana L, Potestio R, Lattanzi G, Siliqi D
|
| RgGuinier |
2.9 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
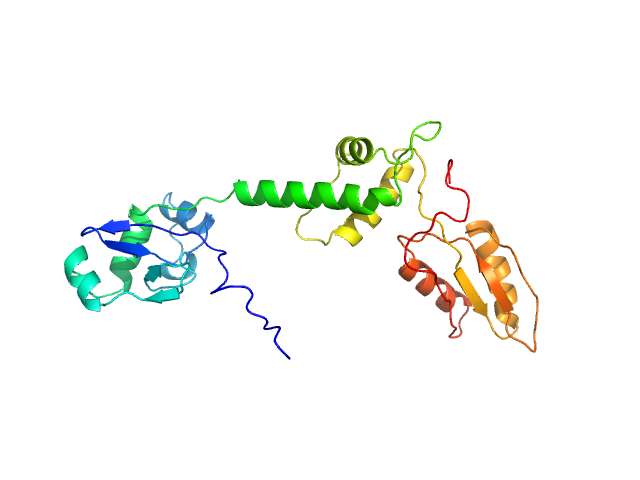
| Sample: |
Ribosome maturation protein SBDS (I167T) monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 6
|
Structural Implications of Missense Point Mutations in Shwachman–Bodian–Diamond Syndrome Protein (SBDS): A Combined SAXS/MD Investigation
ACS Omega (2025)
Mattiotti G, Nanna V, Giulini M, Alberga D, Mangiatordi G, Sánchez-Puig N, Saviano M, Tubiana L, Potestio R, Lattanzi G, Siliqi D
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Ribosome maturation protein SBDS (R175W) monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris pH 8.0, 10% glycerol, 300 mM NaCl, 5 mM MgCl2., pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 6
|
Structural Implications of Missense Point Mutations in Shwachman–Bodian–Diamond Syndrome Protein (SBDS): A Combined SAXS/MD Investigation
ACS Omega (2025)
Mattiotti G, Nanna V, Giulini M, Alberga D, Mangiatordi G, Sánchez-Puig N, Saviano M, Tubiana L, Potestio R, Lattanzi G, Siliqi D
|
| RgGuinier |
3.1 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|
|
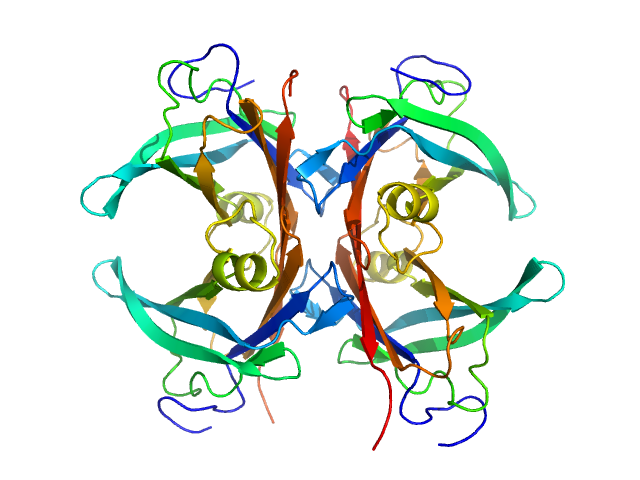
| Sample: |
Transthyretin tetramer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2022 Sep 4
|
Differentiating the solution structures and stability of transthyretin tetramer complexed with tolcapone and tafamidis using SEC-SWAXS and NMR.
J Appl Crystallogr 58(Pt 4):1373-1383 (2025)
Shih O, Feng YC, Agrawal S, Liao KF, Yeh YQ, Chang JW, Yu TY, Jeng US
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
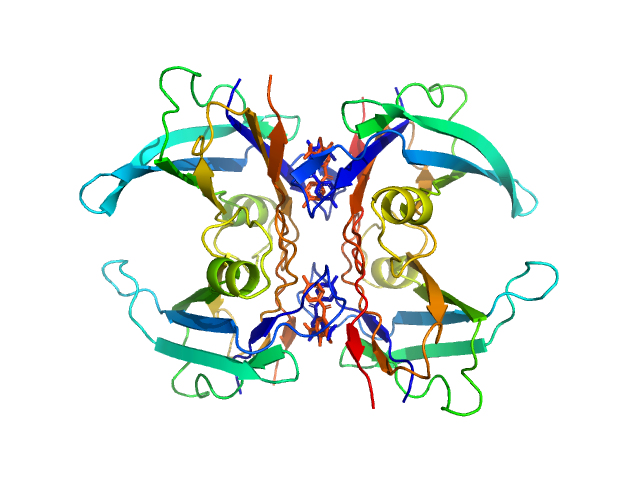
|
| Sample: |
Transthyretin tetramer, 55 kDa Homo sapiens protein
Tolcapone dimer, 1 kDa
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 May 19
|
Differentiating the solution structures and stability of transthyretin tetramer complexed with tolcapone and tafamidis using SEC-SWAXS and NMR.
J Appl Crystallogr 58(Pt 4):1373-1383 (2025)
Shih O, Feng YC, Agrawal S, Liao KF, Yeh YQ, Chang JW, Yu TY, Jeng US
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|
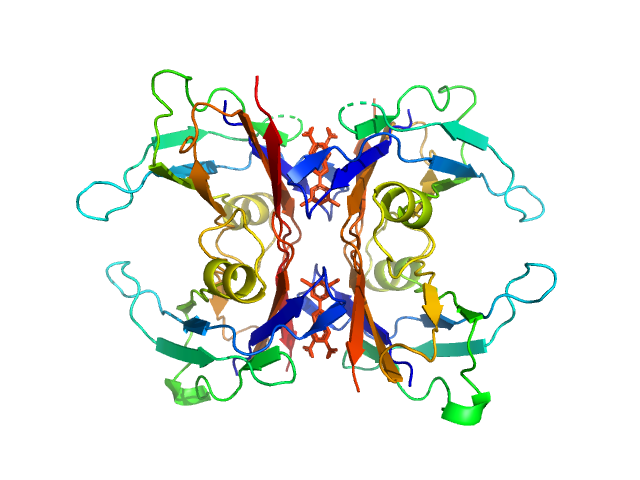
|
| Sample: |
Transthyretin tetramer, 55 kDa Homo sapiens protein
Tafamidis dimer, 1 kDa
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 May 19
|
Differentiating the solution structures and stability of transthyretin tetramer complexed with tolcapone and tafamidis using SEC-SWAXS and NMR.
J Appl Crystallogr 58(Pt 4):1373-1383 (2025)
Shih O, Feng YC, Agrawal S, Liao KF, Yeh YQ, Chang JW, Yu TY, Jeng US
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
69 |
nm3 |
|
|