|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl 42%D2O, pH: 6.6 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2015 Sep 21
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
222 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl 42%D2O, pH: 6.6 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2015 Nov 23
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl 42%D2O, pH: 6.6 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2019 Jul 21
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl 42%D2O, pH: 6.6 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2015 May 23
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
5.1 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
125 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D' monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl, pH: 6.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jun 13
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
4.7 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
346 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase dimer, 52 kDa Pyrococcus furiosus protein
50S ribosomal protein L7Ae dimer, 27 kDa Pyrococcus furiosus protein
NOP5/NOP56 related protein dimer, 94 kDa Pyrococcus furiosus protein
Pyrococcus furiosus sR26 stabilized construct monomer, 24 kDa Pyrococcus furiosus RNA
Pyrococcus furiosus sR26 substrate D monomer, 4 kDa Pyrococcus furiosus RNA
|
| Buffer: |
50 mM phosphate 500 mM NaCl, pH: 6.6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2015 Jul 13
|
The guide sRNA sequence determines the activity level of box C/D RNPs.
Elife 9 (2020)
Graziadei A, Gabel F, Kirkpatrick J, Carlomagno T
|
| RgGuinier |
5.0 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
429 |
nm3 |
|
|
|
|
|
|
|
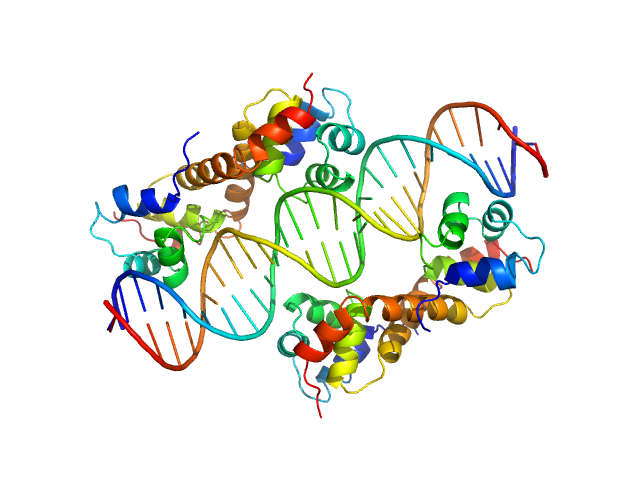
| Sample: |
Uncharacterized protein dimer, 22 kDa Pseudomonas aeruginosa protein
DNA Duplex dimer, 20 kDa DNA
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 5% (v/v) glycerol, and 1 mM PMSF, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Sep 19
|
Structural Insights Into the Transcriptional Regulation of HigBA Toxin–Antitoxin System by Antitoxin HigA in Pseudomonas aeruginosa
Frontiers in Microbiology 10 (2020)
Liu Y, Gao Z, Liu G, Geng Z, Dong Y, Zhang H
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
81 |
nm3 |
|
|
|
|
|
|
|
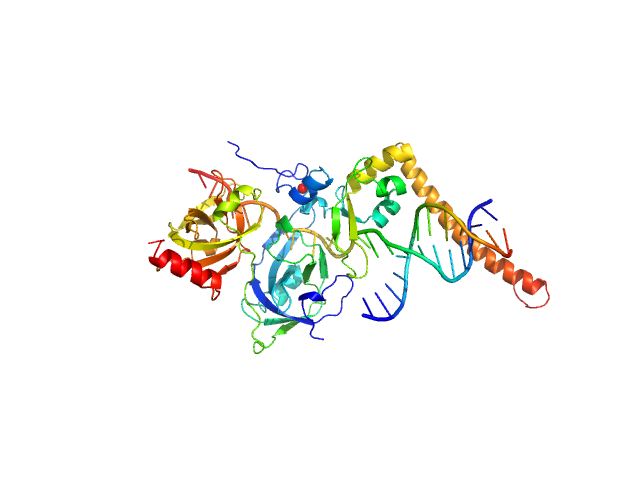
| Sample: |
DNA repair protein complementing XP-A cells monomer, 17 kDa Homo sapiens protein
Replication protein A 70 kDa DNA-binding subunit monomer, 27 kDa Homo sapiens protein
3-prime Nucleotide Excision Repair Junction Model Substrate monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 17 Nov 2
|
A key interaction with RPA orients XPA in NER complexes.
Nucleic Acids Res (2020)
Topolska-Woś AM, Sugitani N, Cordoba JJ, Le Meur KV, Le Meur RA, Kim HS, Yeo JE, Rosenberg D, Hammel M, Schärer OD, Chazin WJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|
|
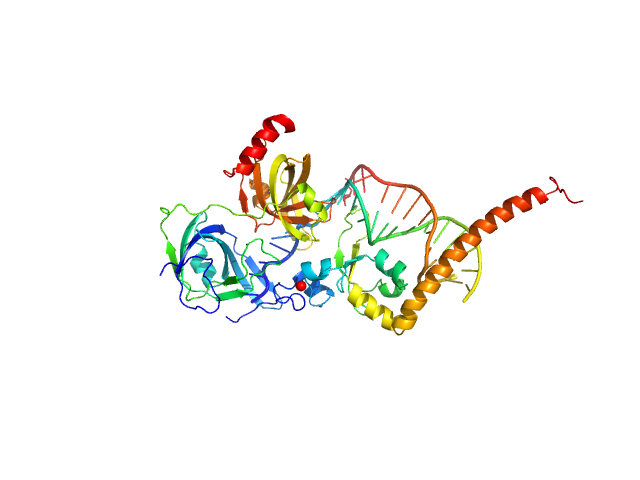
| Sample: |
DNA repair protein complementing XP-A cells monomer, 17 kDa Homo sapiens protein
Replication protein A 70 kDa DNA-binding subunit monomer, 27 kDa Homo sapiens protein
5-prime Nucleotide Excision Repair Junction Model Substrate monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 4
|
A key interaction with RPA orients XPA in NER complexes.
Nucleic Acids Res (2020)
Topolska-Woś AM, Sugitani N, Cordoba JJ, Le Meur KV, Le Meur RA, Kim HS, Yeo JE, Rosenberg D, Hammel M, Schärer OD, Chazin WJ
|
| RgGuinier |
2.9 |
nm |
| Dmax |
97.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
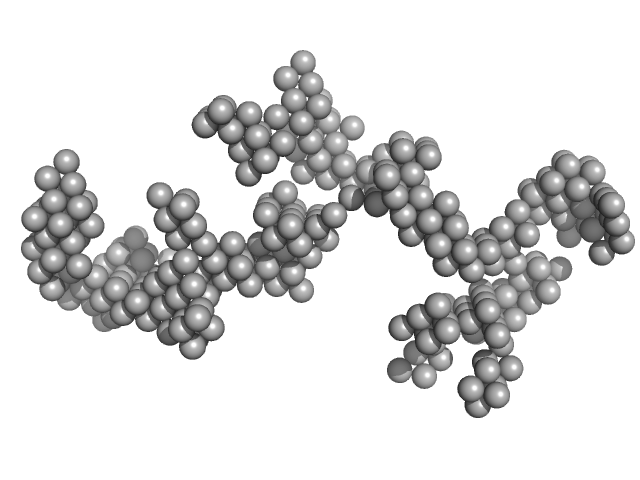
|
| Sample: |
Braveheart RNA monomer, 205 kDa Homo sapiens RNA
Cellular nucleic acid-binding protein monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
9.8 |
nm |
| Dmax |
30.2 |
nm |
| VolumePorod |
1660 |
nm3 |
|
|