|
|
|
|

|
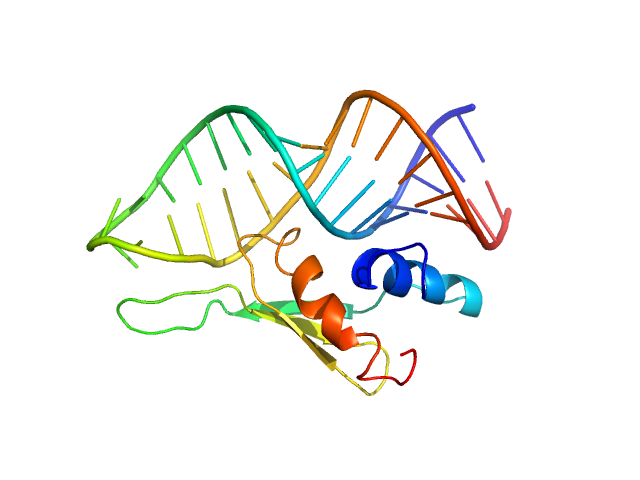
| Sample: |
Cellular nucleic acid-binding protein monomer, 22 kDa Homo sapiens protein
Braveheart Fragment 1 monomer, 116 kDa Homo sapiens RNA
|
| Buffer: |
50 mM HEPES-KOH, 100 mM KCl, 6 mM MgCl2, pH: 7.6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 23
|
Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution
Nature Communications 11(1) (2020)
Kim D, Thiel B, Mrozowich T, Hennelly S, Hofacker I, Patel T, Sanbonmatsu K
|
| RgGuinier |
8.2 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
455 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Double-stranded RNA-binding protein Staufen homolog 1 - RNA binding domain 4 monomer, 8 kDa Homo sapiens protein
ADP-ribosylation factor1 - short monomer, 11 kDa Homo sapiens RNA
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 3.5 mM 2-mercaptoethanol, pH: 6.8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2015 Dec 10
|
Staufen1 reads out structure and sequence features in ARF1 dsRNA for target recognition.
Nucleic Acids Res (2019)
Yadav DK, Zigáčková D, Zlobina M, Klumpler T, Beaumont C, Kubíčková M, Vaňáčová Š, Lukavsky PJ
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
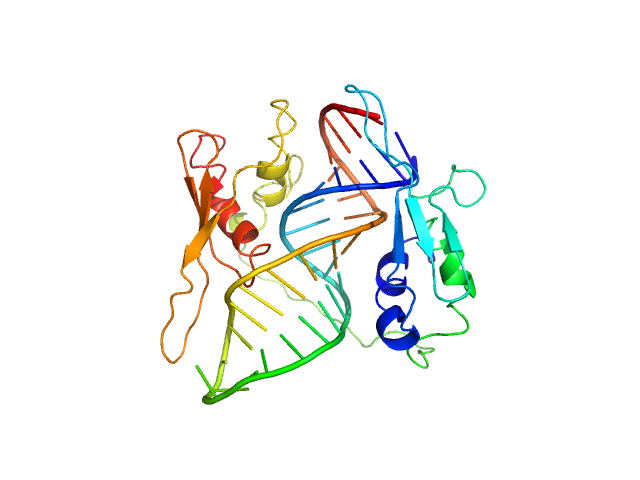
| Sample: |
ADP-ribosylation factor1 - short monomer, 11 kDa Homo sapiens RNA
Double-stranded RNA-binding protein Staufen homolog 1 - RNA binding domain 3 and 4 monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate buffer, 300 mM NaCl, 500 mM imidazole, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2018 May 29
|
Staufen1 reads out structure and sequence features in ARF1 dsRNA for target recognition.
Nucleic Acids Res (2019)
Yadav DK, Zigáčková D, Zlobina M, Klumpler T, Beaumont C, Kubíčková M, Vaňáčová Š, Lukavsky PJ
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Double-stranded RNA-binding protein Staufen homolog 1 - RNA binding domain 3 and 4 monomer, 20 kDa Homo sapiens protein
ADP-ribosylation factor1 - long monomer, 16 kDa Homo sapiens RNA
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 3.5 mM 2-mercaptoethanol, pH: 6.8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Sep 20
|
Staufen1 reads out structure and sequence features in ARF1 dsRNA for target recognition.
Nucleic Acids Res (2019)
Yadav DK, Zigáčková D, Zlobina M, Klumpler T, Beaumont C, Kubíčková M, Vaňáčová Š, Lukavsky PJ
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
|
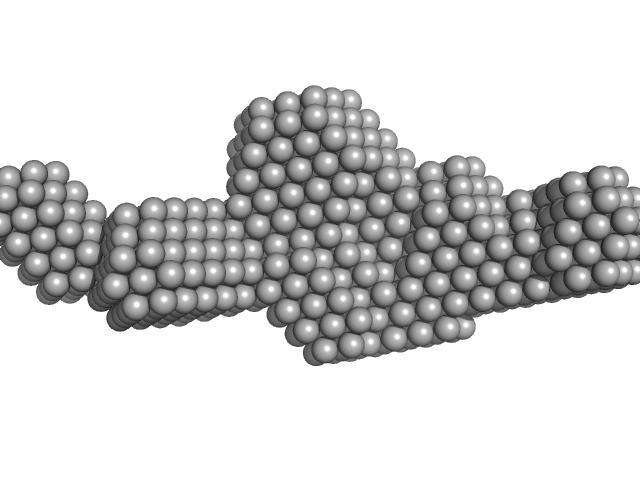
| Sample: |
Sa0446 binding sequence 40bp monomer, 25 kDa DNA
Transcriptional regulator Lrs14-like protein dimer, 33 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
300 mM NaCl, 20 mM HEPES, pH 7.5, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 5
|
Solution Structure of Archaeal Biofilm Regulator 2 (AbfR2) in Complex with 40 bp DNA
Marian Vogt
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
J-DNA binding domain monomer, 21 kDa Leishmania tarentolae protein
J-DNA (23mer) monomer, 14 kDa DNA
|
| Buffer: |
20 mM HEPES, 200 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 21
|
The domain architecture of protozoan protein J-DNA-binding protein 1 suggests synergy between base J DNA binding and thymidine hydroxylase activity.
J Biol Chem (2019)
Adamopoulos A, Heidebrecht T, Roosendaal J, Touw WG, Phan IQ, Beijnen J, Perrakis A
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
43 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Thymine dioxygenase JBP1 monomer, 93 kDa Leishmania tarentolae protein
J-DNA (23mer) monomer, 14 kDa DNA
|
| Buffer: |
20 mM HEPES, 200 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 21
|
The domain architecture of protozoan protein J-DNA-binding protein 1 suggests synergy between base J DNA binding and thymidine hydroxylase activity.
J Biol Chem (2019)
Adamopoulos A, Heidebrecht T, Roosendaal J, Touw WG, Phan IQ, Beijnen J, Perrakis A
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
148 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Dosage compensation regulator monomer, 29 kDa Drosophila melanogaster protein
RoX2 stem-loop 7, 18-mer fragment monomer, 12 kDa synthetic construct RNA
|
| Buffer: |
20 mM NaPO4, 200 mM NaCl, 1 mM DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 29
|
Structure, dynamics and roX2-lncRNA binding of tandem double-stranded RNA binding domains dsRBD1,2 of Drosophila helicase Maleless.
Nucleic Acids Res 47(8):4319-4333 (2019)
Ankush Jagtap PK, Müller M, Masiewicz P, von Bülow S, Hollmann NM, Chen PC, Simon B, Thomae AW, Becker PB, Hennig J
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Histone H3 dimer, 31 kDa Xenopus laevis protein
Histone H4 dimer, 22 kDa Xenopus laevis protein
Histone H2a dimer, 28 kDa Xenopus laevis protein
Histone H2b dimer, 27 kDa Xenopus laevis protein
147bp 601 Widom sequence monomer, 45 kDa synthetic construct DNA
|
| Buffer: |
15 mM HEPES, 200 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 11
|
A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep 27(2):387-399.e7 (2019)
Marabelli C, Marrocco B, Pilotto S, Chittori S, Picaud S, Marchese S, Ciossani G, Forneris F, Filippakopoulos P, Schoehn G, Rhodes D, Subramaniam S, Mattevi A
|
| RgGuinier |
4.4 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
353 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lysyne-specific Demethylase LSD2 monomer, 89 kDa Homo sapiens protein
NPAC linker+DH (delta-205) tetramer, 150 kDa Homo sapiens protein
Histone H3 dimer, 31 kDa Xenopus laevis protein
Histone H4 dimer, 22 kDa Xenopus laevis protein
Histone H2a dimer, 28 kDa Xenopus laevis protein
Histone H2b dimer, 27 kDa Xenopus laevis protein
147bp 601 Widom sequence monomer, 45 kDa synthetic construct DNA
|
| Buffer: |
15 mM HEPES, 200 mM NaCl, pH: 7.3 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Dec 11
|
A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep 27(2):387-399.e7 (2019)
Marabelli C, Marrocco B, Pilotto S, Chittori S, Picaud S, Marchese S, Ciossani G, Forneris F, Filippakopoulos P, Schoehn G, Rhodes D, Subramaniam S, Mattevi A
|
| RgGuinier |
7.8 |
nm |
| Dmax |
30.4 |
nm |
| VolumePorod |
1100 |
nm3 |
|
|