|
|
|
|
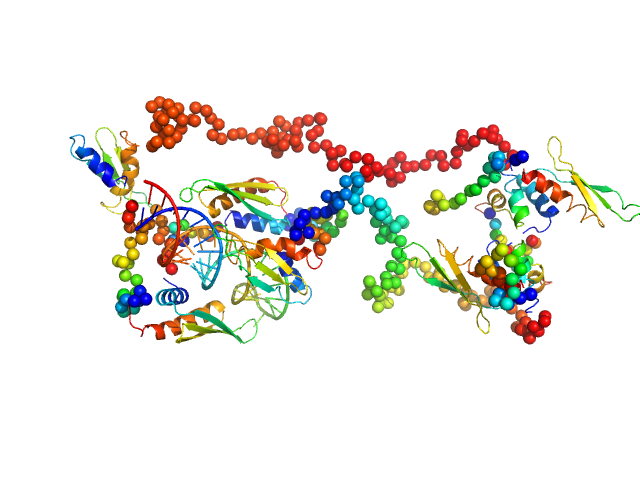
|
| Sample: |
Double-stranded RNA-binding protein Staufen homolog 1 (Δ1-177) dimer, 89 kDa Homo sapiens protein
3'UTR fragment of ADP-ribosylation factor 1 monomer, 14 kDa Homo sapiens RNA
|
| Buffer: |
50 mM TRIS, 300 mM NaCl, 3.8 mM β-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2020 May 4
|
A Simple Protocol for Visualization of RNA-Protein Complexes by Atomic Force Microscopy.
Curr Protoc 5(1):e70084 (2025)
Tripepi A, Shakoor H, Klapetek P
|
| RgGuinier |
4.9 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
129 |
nm3 |
|
|
|
|
|
|
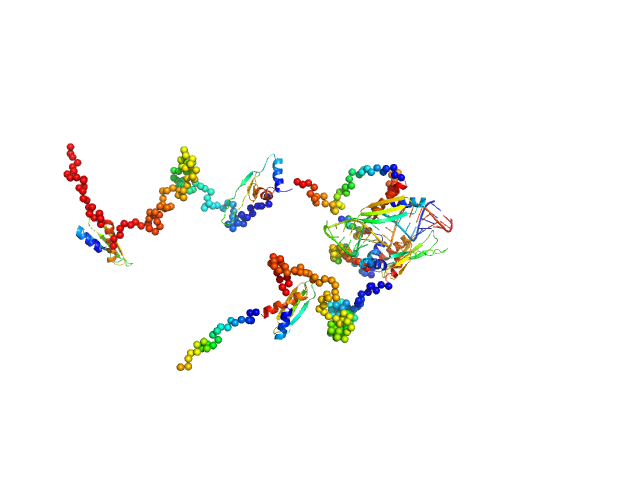
|
| Sample: |
3'UTR fragment of ADP-ribosylation factor 1 monomer, 14 kDa Homo sapiens RNA
Double-stranded RNA-binding protein Staufen homolog 1 with truncated RNA-binding domain 2 and truncated Staufen-swapping (ΔSSM) dimer, 81 kDa Homo sapiens protein
|
| Buffer: |
50 mM TRIS, 300 mM NaCl, 3.8 mM β-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2024 Jan 12
|
A Simple Protocol for Visualization of RNA-Protein Complexes by Atomic Force Microscopy.
Curr Protoc 5(1):e70084 (2025)
Tripepi A, Shakoor H, Klapetek P
|
| RgGuinier |
5.5 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
139 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
E3 ubiquitin/ISG15 ligase TRIM25 dimer, 100 kDa Homo sapiens protein
Pre-let-7-a-1@1 monomer, 9 kDa synthetic construct RNA
|
| Buffer: |
20 mM MES, 75 mM NaCl, 1 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 14
|
The molecular dissection of TRIM25's RNA-binding mechanism provides key insights into its antiviral activity.
Nat Commun 15(1):8485 (2024)
Álvarez L, Haubrich K, Iselin L, Gillioz L, Ruscica V, Lapouge K, Augsten S, Huppertz I, Choudhury NR, Simon B, Masiewicz P, Lethier M, Cusack S, Rittinger K, Gabel F, Leitner A, Michlewski G, Hentze MW, Allain FHT, Castello A, Hennig J
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
E3 ubiquitin/ISG15 ligase TRIM25 dimer, 100 kDa Homo sapiens protein
Pre-let-7-a-1@1 monomer, 9 kDa synthetic construct RNA
|
| Buffer: |
20 mM MES, 75 mM NaCl, 1 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jun 4
|
The molecular dissection of TRIM25's RNA-binding mechanism provides key insights into its antiviral activity.
Nat Commun 15(1):8485 (2024)
Álvarez L, Haubrich K, Iselin L, Gillioz L, Ruscica V, Lapouge K, Augsten S, Huppertz I, Choudhury NR, Simon B, Masiewicz P, Lethier M, Cusack S, Rittinger K, Gabel F, Leitner A, Michlewski G, Hentze MW, Allain FHT, Castello A, Hennig J
|
| RgGuinier |
5.7 |
nm |
| Dmax |
23.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
E3 ubiquitin/ISG15 ligase TRIM25 dimer, 100 kDa Homo sapiens protein
Lnczc3h7a_304-326 monomer, 8 kDa Homo sapiens RNA
|
| Buffer: |
20 mM MES, 75 mM NaCl, 1 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 12
|
The molecular dissection of TRIM25's RNA-binding mechanism provides key insights into its antiviral activity.
Nat Commun 15(1):8485 (2024)
Álvarez L, Haubrich K, Iselin L, Gillioz L, Ruscica V, Lapouge K, Augsten S, Huppertz I, Choudhury NR, Simon B, Masiewicz P, Lethier M, Cusack S, Rittinger K, Gabel F, Leitner A, Michlewski G, Hentze MW, Allain FHT, Castello A, Hennig J
|
| RgGuinier |
5.8 |
nm |
| Dmax |
16.6 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDV54_fit1_model1.png)
|
| Sample: |
Phage antirepressor protein Cro dimer, 24 kDa Escherichia coli O157:H7 protein
IR1 22 base pair dsDNA monomer, 14 kDa DNA
|
| Buffer: |
20 mM sodium acetate, 150 mM NaCl, 1 mM TCEP, pH: 5.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Apr 13
|
Structural-function relationship of YdaS, a Cro-type repressor in the cryptic prophage CP-933P from Escherichia coli O157:H7
Marusa Prolic Kalinsek
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
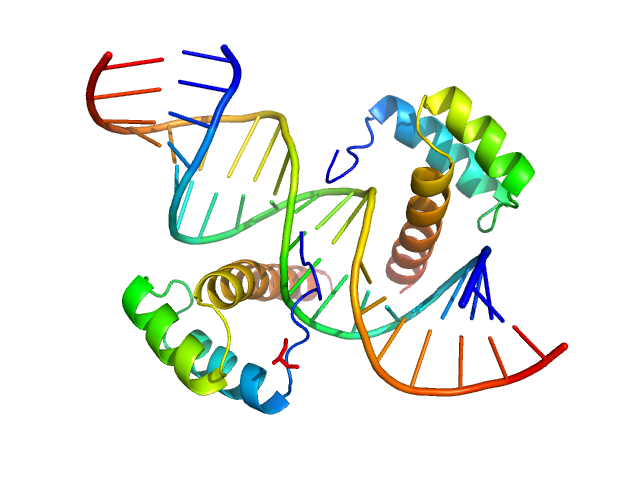
|
| Sample: |
Cone-rod homeobox protein, 20 kDa Homo sapiens protein
Ret4 sense strand, 6 kDa synthetic construct DNA
Ret4 antisense strand, 6 kDa synthetic construct DNA
|
| Buffer: |
20 mM Tris, 150 mM KCl, 5 % glycerol, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Mar 25
|
Molecular basis of CRX/DNA recognition and stoichiometry at the Ret4 response element
Structure (2024)
Srivastava D, Gowribidanur-Chinnaswamy P, Gaur P, Spies M, Swaroop A, Artemyev N
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Antitoxin HigA-2 dimer, 23 kDa Vibrio cholerae serotype … protein
DNA operator dimer, 20 kDa DNA
|
| Buffer: |
20 mM Tris pH 8, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Apr 6
|
Fuzzy recognition by the prokaryotic transcription factor HigA2 from Vibrio cholerae.
Nat Commun 15(1):3105 (2024)
Hadži S, Živič Z, Kovačič M, Zavrtanik U, Haeserts S, Charlier D, Plavec J, Volkov AN, Lah J, Loris R
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|
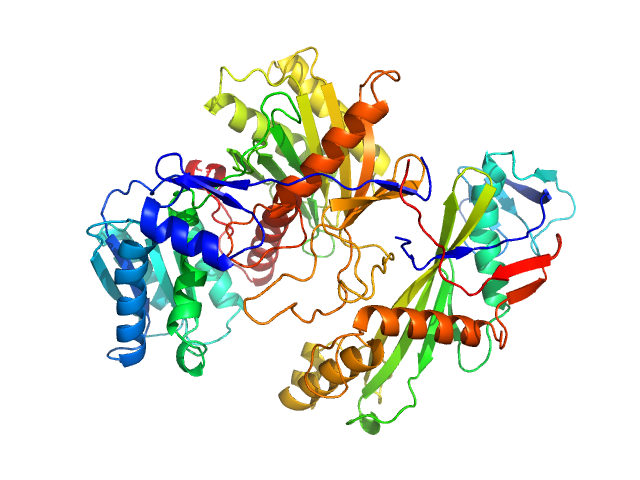
|
| Sample: |
Piwi protein AF_1318 (Archaeoglobus fulgidus AfAgo protein) monomer, 51 kDa Archaeoglobus fulgidus (strain … protein
Uncharacterized protein (AfAgo-N protein containing N-L1-L2 domains) monomer, 31 kDa Archaeoglobus fulgidus DSM … protein
5'-end phosphorylated DNA oligoduplex, 14 bp (MZ1288) monomer, 9 kDa DNA
|
| Buffer: |
20 mM TrisHCl pH 7.5, 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 6
|
The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res (2024)
Manakova E, Golovinas E, Pocevičiūtė R, Sasnauskas G, Silanskas A, Rutkauskas D, Jankunec M, Zagorskaitė E, Jurgelaitis E, Grybauskas A, Venclovas Č, Zaremba M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Single-chain full Archaeoglobus fulgidus Argonaute monomer, 78 kDa Archaeoglobus fulgidus protein
5'-end phosphorylated DNA oligoduplex, 14 bp (MZ1288) monomer, 9 kDa DNA
|
| Buffer: |
20 mM TrisHCl pH7.5, 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 6
|
The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res (2024)
Manakova E, Golovinas E, Pocevičiūtė R, Sasnauskas G, Silanskas A, Rutkauskas D, Jankunec M, Zagorskaitė E, Jurgelaitis E, Grybauskas A, Venclovas Č, Zaremba M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
131 |
nm3 |
|
|