|
|
|
|
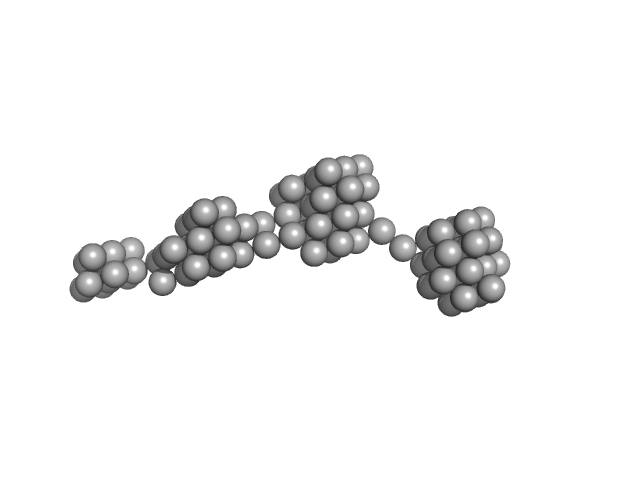
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 23
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
12.4 |
nm |
| Dmax |
59.0 |
nm |
| VolumePorod |
1820 |
nm3 |
|
|
|
|
|
|
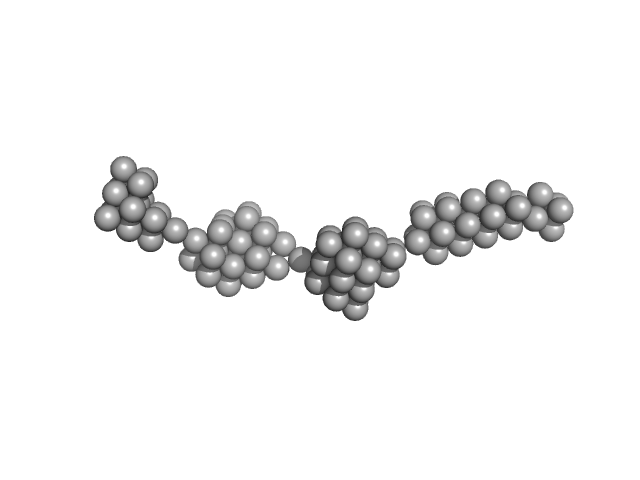
|
| Sample: |
Persulfide dioxygenase ETHE1, mitochondrial dimer, 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 2 mM TCEP, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 23
|
Distinctive features and structural significance of the Homo sapiens ethylmalonic encephalopathy protein iron binding site
Al Kikhney,
Marco Salomone-Stagni
|
| RgGuinier |
14.2 |
nm |
| Dmax |
63.5 |
nm |
| VolumePorod |
2317 |
nm3 |
|
|
|
|
|
|
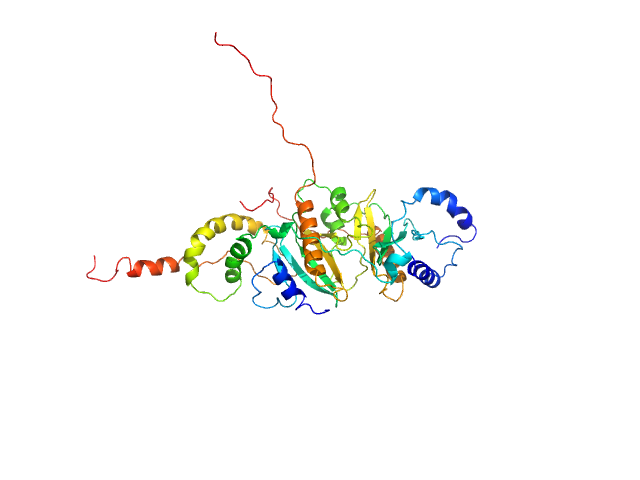
|
| Sample: |
SycH putative yopH targeting protein dimer, 32 kDa Yersinia pseudotuberculosis protein
Tyrosine-protein phosphatase YopH monomer, 14 kDa Yersinia pseudotuberculosis protein
|
| Buffer: |
50 mM HEPES 2mM TCEP, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Aug 1
|
Global Disordering in Stereo-Specific Protein Association
Biophysical Journal 112(3):33a (2017)
Gupta A, Reinartz I, Spilotros A, Jonna V, Hofer A, Svergun D, Schug A, Wolf-Watz M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
|
|
|
|
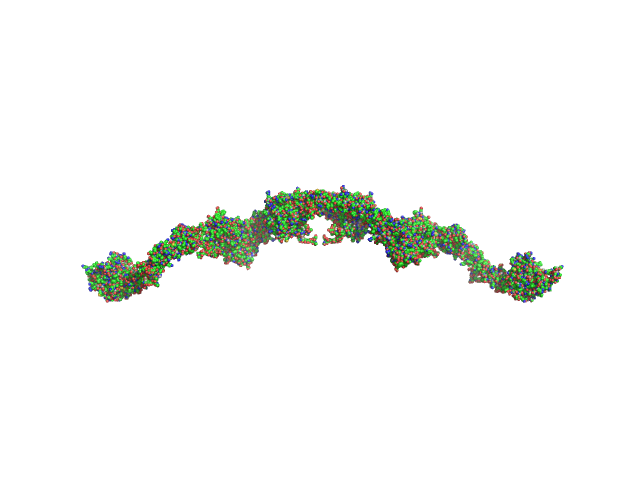
|
| Sample: |
Complement C1r subcomponent dimer, 156 kDa Homo sapiens protein
Complement C1s subcomponent dimer, 150 kDa Homo sapiens protein
|
| Buffer: |
50 mM TrisHCl, 145 mM NaCl, 3 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Dec 8
|
Structure and activation of C1, the complex initiating the classical pathway of the complement cascade.
Proc Natl Acad Sci U S A 114(5):986-991 (2017)
Mortensen SA, Sander B, Jensen RK, Pedersen JS, Golas MM, Jensenius JC, Hansen AG, Thiel S, Andersen GR
|
|
|
|
|
|
|
|
| Sample: |
Complement C1q subcomponent subunit C hexamer, 142 kDa Homo sapiens protein
Complement C1q subcomponent subunit B hexamer, 142 kDa Homo sapiens protein
Complement C1q subcomponent subunit A hexamer, 142 kDa Homo sapiens protein
|
| Buffer: |
50 mM TrisHCl, 145 mM NaCl, 3 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Dec 8
|
Structure and activation of C1, the complex initiating the classical pathway of the complement cascade.
Proc Natl Acad Sci U S A 114(5):986-991 (2017)
Mortensen SA, Sander B, Jensen RK, Pedersen JS, Golas MM, Jensenius JC, Hansen AG, Thiel S, Andersen GR
|
|
|
|
|
|
|

|
| Sample: |
Complement C1q subcomponent subunit C hexamer, 142 kDa Homo sapiens protein
Complement C1q subcomponent subunit B hexamer, 142 kDa Homo sapiens protein
Complement C1q subcomponent subunit A hexamer, 142 kDa Homo sapiens protein
Complement C1r subcomponent dimer, 156 kDa Homo sapiens protein
Complement C1s subcomponent dimer, 150 kDa Homo sapiens protein
|
| Buffer: |
50 mM EPPS, 145 mM NaCl, 3 mM CaCl2, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Aug 16
|
Structure and activation of C1, the complex initiating the classical pathway of the complement cascade.
Proc Natl Acad Sci U S A 114(5):986-991 (2017)
Mortensen SA, Sander B, Jensen RK, Pedersen JS, Golas MM, Jensenius JC, Hansen AG, Thiel S, Andersen GR
|
| RgGuinier |
11.5 |
nm |
| Dmax |
36.6 |
nm |
|
|
|
|
|
|

|
| Sample: |
Iron-regulated outer membrane lipoprotein FrpD monomer, 27 kDa Neisseria meningitidis protein
|
| Buffer: |
10 mM Tris-HCl 150 mM NaCl 0.01% NaN3, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Oct 19
|
Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep 7:40408 (2017)
Sviridova E, Rezacova P, Bondar A, Veverka V, Novak P, Schenk G, Svergun DI, Kuta Smatanova I, Bumba L
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-regulated outer membrane lipoprotein FrpD monomer, 27 kDa Neisseria meningitidis protein
Iron-regulated protein FrpC monomer, 46 kDa Neisseria meningitidis protein
|
| Buffer: |
50 mM Tris-HCl 150 mM NaCl 0.01% NaN3, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Oct 19
|
Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep 7:40408 (2017)
Sviridova E, Rezacova P, Bondar A, Veverka V, Novak P, Schenk G, Svergun DI, Kuta Smatanova I, Bumba L
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
39 kDa FK506-binding nuclear protein tetramer, 163 kDa Drosophila melanogaster protein
|
| Buffer: |
50 mM Na2HPO4 150 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2014 Nov 14
|
Nucleoplasmin-like domain of FKBP39 from Drosophila melanogaster forms a tetramer with partly disordered tentacle-like C-terminal segments.
Sci Rep 7:40405 (2017)
Kozłowska M, Tarczewska A, Jakób M, Bystranowska D, Taube M, Kozak M, Czarnocki-Cieciura M, Dziembowski A, Orłowski M, Tkocz K, Ożyhar A
|
| RgGuinier |
5.7 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
275 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Outer membrane protein IcsA (53-758) monomer, 72 kDa Shigella flexneri protein
|
| Buffer: |
50 mM Tris 150 mM NaCl 10 mM CaCl2 3% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 May 18
|
The Shigella Virulence Factor IcsA Relieves N-WASP Autoinhibition by Displacing the Verprolin Homology/Cofilin/Acidic (VCA) Domain.
J Biol Chem 292(1):134-145 (2017)
Mauricio RP, Jeffries CM, Svergun DI, Deane JE
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
103 |
nm3 |
|
|