|
|
|
|
|
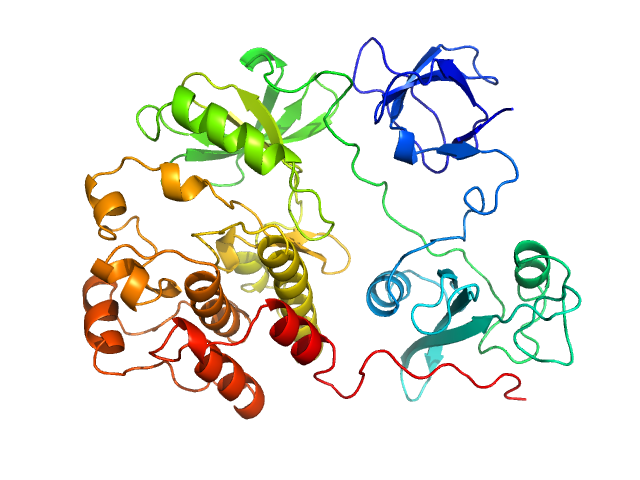
| Sample: |
Proto-oncogene tyrosine-protein kinase Src monomer, 52 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8.0, 150 mM NaCl, 1% glycerol and 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 8
|
An allosteric switch between the activation loop and a c-terminal palindromic phospho-motif controls c-Src function
Nature Communications 14(1) (2023)
Cuesta-Hernández H, Contreras J, Soriano-Maldonado P, Sánchez-Wandelmer J, Yeung W, Martín-Hurtado A, Muñoz I, Kannan N, Llimargas M, Muñoz J, Plaza-Menacho I
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Hepatocyte nuclear factor 1-alpha monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 1 mM TCEP,, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 25
|
Structural properties of the HNF-1A transactivation domain
Frontiers in Molecular Biosciences 10 (2023)
Kind L, Driver M, Raasakka A, Onck P, Njølstad P, Arnesen T, Kursula P
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Hepatocyte nuclear factor 1-alpha monomer, 62 kDa Homo sapiens protein
|
| Buffer: |
4 mM Tris, 100 mM NaCl, 1 mM TCEP,, pH: 8.5 |
| Experiment: |
SAXS
data collected at CoSAXS, MAX IV on 2022 Apr 2
|
Structural properties of the HNF-1A transactivation domain
Frontiers in Molecular Biosciences 10 (2023)
Kind L, Driver M, Raasakka A, Onck P, Njølstad P, Arnesen T, Kursula P
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
109 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
HOTag-GS-Ubiquitin tetramer, 51 kDa synthetic construct protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 16
|
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units
Proceedings of the National Academy of Sciences 120(42) (2023)
Galagedera S, Dao T, Enos S, Chaudhuri A, Schmit J, Castañeda C
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
HOTag6-(GS)2-Ubiquitin tetramer, 52 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Feb 21
|
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units
Proceedings of the National Academy of Sciences 120(42) (2023)
Galagedera S, Dao T, Enos S, Chaudhuri A, Schmit J, Castañeda C
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
HOTag-(GS)4-Ubiquitin tetramer, 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 16
|
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units
Proceedings of the National Academy of Sciences 120(42) (2023)
Galagedera S, Dao T, Enos S, Chaudhuri A, Schmit J, Castañeda C
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
HOTag-(GS)10-Ubiquitin tetramer, 56 kDa synthetic construct protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 16
|
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units
Proceedings of the National Academy of Sciences 120(42) (2023)
Galagedera S, Dao T, Enos S, Chaudhuri A, Schmit J, Castañeda C
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
HOTag6-(GS)25-Ubiquitin tetramer, 65 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Feb 21
|
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units
Proceedings of the National Academy of Sciences 120(42) (2023)
Galagedera S, Dao T, Enos S, Chaudhuri A, Schmit J, Castañeda C
|
| RgGuinier |
4.7 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
HOTag-(GS)50-Ubiquitin tetramer, 80 kDa synthetic construct protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 16
|
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units
Proceedings of the National Academy of Sciences 120(42) (2023)
Galagedera S, Dao T, Enos S, Chaudhuri A, Schmit J, Castañeda C
|
| RgGuinier |
5.6 |
nm |
| Dmax |
22.7 |
nm |
| VolumePorod |
193 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
HOTag-PA-Ubiquitin tetramer, 51 kDa synthetic construct protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 16
|
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units
Proceedings of the National Academy of Sciences 120(42) (2023)
Galagedera S, Dao T, Enos S, Chaudhuri A, Schmit J, Castañeda C
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
73 |
nm3 |
|
|