|
|
|
|
|
| Sample: |
Human metaphase chromosomes in 5 mM magnesium chloride monomer, 0 kDa Homo sapiens
|
| Buffer: |
10 mM PIPES, 5 mM magnesium chloride, 10 mM sodium chloride, 120 mM potassium chloride, 40% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2013 Jun 6
|
Frozen-hydrated chromatin from metaphase chromosomes has an interdigitated multilayer structure.
EMBO J 38(7) (2019)
Chicano A, Crosas E, Otón J, Melero R, Engel BD, Daban JR
|
|
|
|
|
|
|
|
| Sample: |
Human metaphase chromosomes in 1 mM hexaamminecobalt(III) chloride monomer, 0 kDa Homo sapiens
|
| Buffer: |
10 mM PIPES, hexaamminecobalt(III) chloride, 40% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at BL11 - NCD, ALBA on 2013 Sep 12
|
Frozen-hydrated chromatin from metaphase chromosomes has an interdigitated multilayer structure.
EMBO J 38(7) (2019)
Chicano A, Crosas E, Otón J, Melero R, Engel BD, Daban JR
|
|
|
|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
Urokinase-type plasminogen activator (Amino Terminal Fragment) monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
20 mM PBS, 5 %(v/v) glycerol, 50 mM NaSO4,, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 18
|
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
J Biol Chem (2019)
Leth JM, Mertens HDT, Leth-Espensen KZ, Jørgensen TJD, Ploug M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
20 mM PBS, 5 %(v/v) glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 1
|
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
J Biol Chem (2019)
Leth JM, Mertens HDT, Leth-Espensen KZ, Jørgensen TJD, Ploug M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
Urokinase-type plasminogen activator (Amino Terminal Fragment) monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
20 mM PBS, 5 %(v/v) glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 1
|
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
J Biol Chem (2019)
Leth JM, Mertens HDT, Leth-Espensen KZ, Jørgensen TJD, Ploug M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
20 mM PBS, 5 %(v/v) glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 5
|
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
J Biol Chem (2019)
Leth JM, Mertens HDT, Leth-Espensen KZ, Jørgensen TJD, Ploug M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
Urokinase-type plasminogen activator (Amino Terminal Fragment) monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
20 mM PBS, 5 %(v/v) glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 5
|
Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond?
J Biol Chem (2019)
Leth JM, Mertens HDT, Leth-Espensen KZ, Jørgensen TJD, Ploug M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cyclic GMP-AMP synthase monomer, 61 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 25
|
cGAS facilitates sensing of extracellular cyclic dinucleotides to activate innate immunity.
EMBO Rep (2019)
Liu H, Moura-Alves P, Pei G, Mollenkopf HJ, Hurwitz R, Wu X, Wang F, Liu S, Ma M, Fei Y, Zhu C, Koehler AB, Oberbeck-Mueller D, Hahnke K, Klemm M, Guhlich-Bornhof U, Ge B, Tuukkanen A, Kolbe M, Dorhoi A, Kaufmann SH
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cyclic GMP-AMP synthase dimer, 123 kDa Homo sapiens protein
2'-O,5'-O-((adenosine-3'-O,5'-O-diyl)bisphosphinico)guanosine dimer, 1 kDa
|
| Buffer: |
20 mM HEPES, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Apr 25
|
cGAS facilitates sensing of extracellular cyclic dinucleotides to activate innate immunity.
EMBO Rep (2019)
Liu H, Moura-Alves P, Pei G, Mollenkopf HJ, Hurwitz R, Wu X, Wang F, Liu S, Ma M, Fei Y, Zhu C, Koehler AB, Oberbeck-Mueller D, Hahnke K, Klemm M, Guhlich-Bornhof U, Ge B, Tuukkanen A, Kolbe M, Dorhoi A, Kaufmann SH
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
|
|
|
|
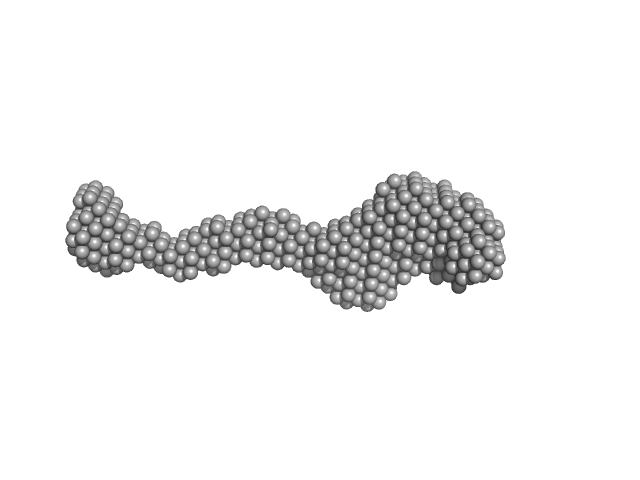
|
| Sample: |
Lysine-specific demethylase 5B monomer, 176 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 300 mM NaCl, 5% (v/v) glycerol, 1mM DTT, pH: 7.7 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with GeniX3D, University of Copenhagen, Department of Drug Design and Pharmacology on 2018 Oct 24
|
Molecular architecture of the Jumonji C family histone demethylase KDM5B.
Sci Rep 9(1):4019 (2019)
Dorosz J, Kristensen LH, Aduri NG, Mirza O, Lousen R, Bucciarelli S, Mehta V, Sellés-Baiget S, Solbak SMØ, Bach A, Mesa P, Hernandez PA, Montoya G, Nguyen TTTN, Rand KD, Boesen T, Gajhede M
|
| RgGuinier |
8.8 |
nm |
| Dmax |
26.9 |
nm |
|
|