|
|
|
|
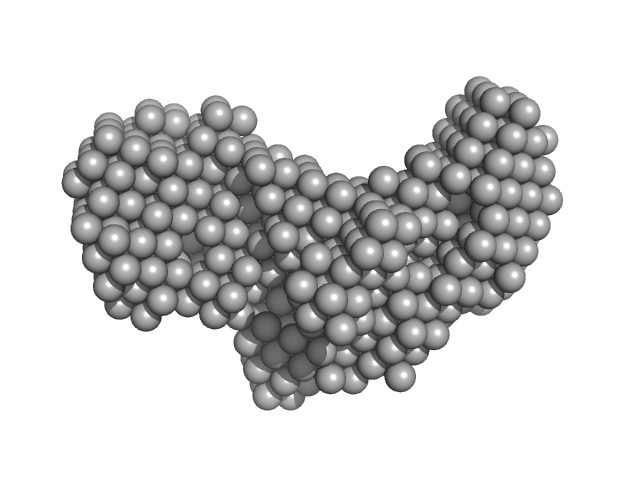
|
| Sample: |
Heat shock 70 kDa protein 1A monomer, 70 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris HCl, 50 mM NaCl, 5 mM Sodium Phosphate, 5 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2018 May 18
|
Structural, thermodynamic and functional studies of human 71 kDa heat shock cognate protein (HSPA8/hHsc70)
Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics :140719 (2021)
Silva N, de Camargo Rodrigues L, Dores-Silva P, Montanari C, Ramos C, Barbosa L, Borges J
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mothers against decapentaplegic homolog 2 (C-terminus phosphomimetic mutant), 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 4
|
Conformational landscape of multidomain SMAD proteins
Computational and Structural Biotechnology Journal (2021)
Gomes T, Martin-Malpartida P, Ruiz L, Aragón E, Cordeiro T, Macias M
|
| RgGuinier |
5.1 |
nm |
| Dmax |
20.0 |
nm |
|
|
|
|
|
|
|
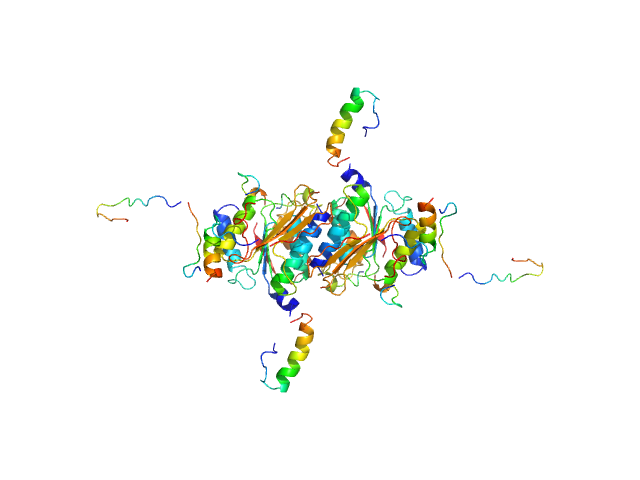
| Sample: |
Mothers against decapentaplegic homolog 2 (C-terminus phosphomimetic mutant), 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 4
|
Conformational landscape of multidomain SMAD proteins
Computational and Structural Biotechnology Journal (2021)
Gomes T, Martin-Malpartida P, Ruiz L, Aragón E, Cordeiro T, Macias M
|
| RgGuinier |
5.2 |
nm |
| Dmax |
20.0 |
nm |
|
|
|
|
|
|
|
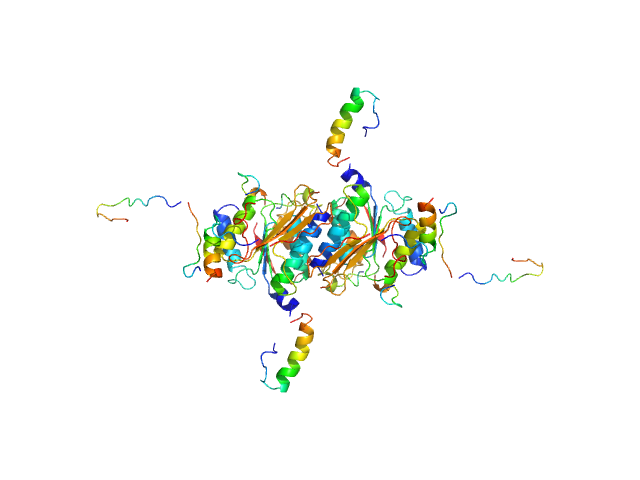
| Sample: |
Mothers against decapentaplegic homolog 2 (C-terminus phosphomimetic mutant), 53 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Apr 4
|
Conformational landscape of multidomain SMAD proteins
Computational and Structural Biotechnology Journal (2021)
Gomes T, Martin-Malpartida P, Ruiz L, Aragón E, Cordeiro T, Macias M
|
| RgGuinier |
5.3 |
nm |
| Dmax |
20.0 |
nm |
|
|
|
|
|
|
|
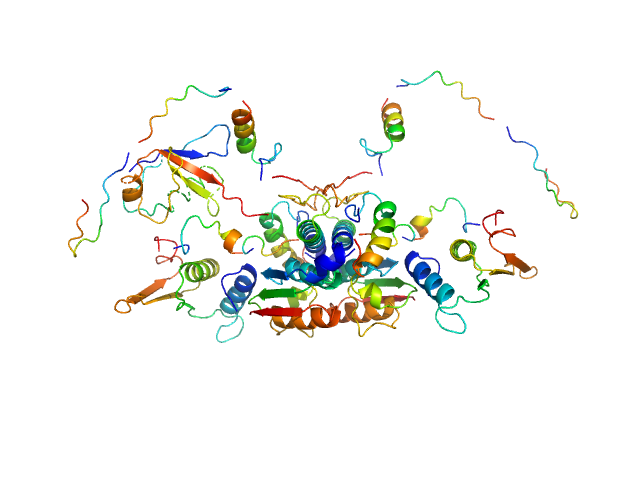
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
Ubiquitin fold modifer 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 29
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
Ubiquitin fold modifer 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 29
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
Ubiquitin fold modifer 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 29
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.0 |
nm |
|
|
|
|
|
|
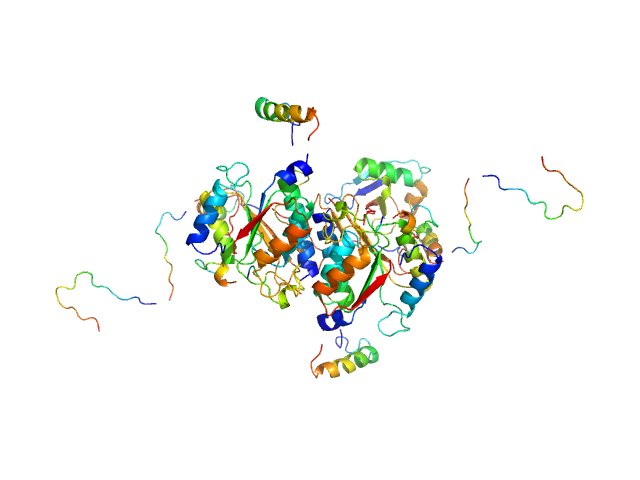
|
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
Ubiquitin fold modifer 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 29
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
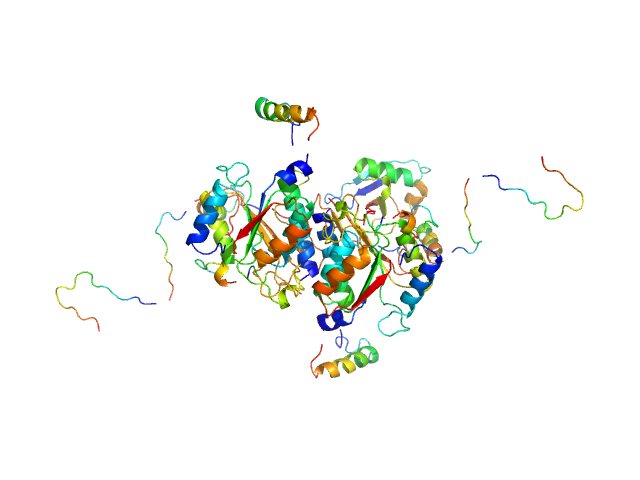
|
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
Ubiquitin fold modifer 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 29
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|
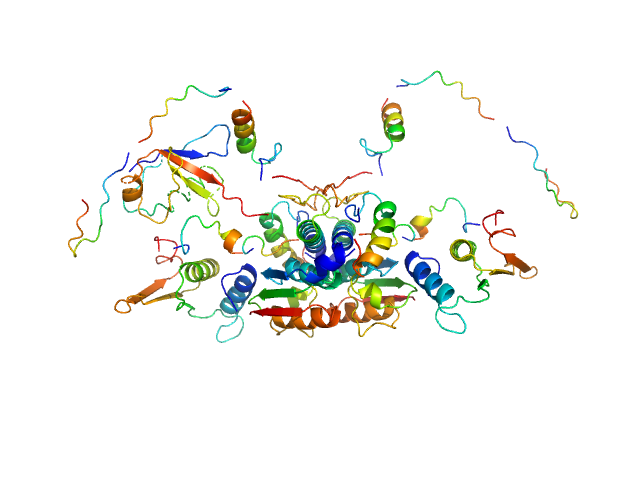
|
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
Ubiquitin fold modifer 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 29
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
104 |
nm3 |
|
|