|
|
|
|
|
| Sample: |
Survival of motor neuron protein-interacting protein yip11 (Gemin2 ΔN80) monomer, 18 kDa Schizosaccharomyces pombe (strain … protein
Survival motor neuron-like protein 1 (Δ36-119) monomer, 8 kDa Schizosaccharomyces pombe protein
|
| Buffer: |
150 mM NaCl, 20 mM HEPES, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 30
|
Identification and structural analysis of the Schizosaccharomyces pombe
SMN complex
Nucleic Acids Research (2021)
Veepaschit J, Viswanathan A, Bordonné R, Grimm C, Fischer U
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Survival of motor neuron protein-interacting protein yip11 (Gemin2 ΔN80) monomer, 18 kDa Schizosaccharomyces pombe (strain … protein
Survival motor neuron-like protein 1 (Δ36-119) monomer, 8 kDa Schizosaccharomyces pombe protein
|
| Buffer: |
150 mM NaCl, 20 mM HEPES, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 29
|
Identification and structural analysis of the Schizosaccharomyces pombe
SMN complex
Nucleic Acids Research (2021)
Veepaschit J, Viswanathan A, Bordonné R, Grimm C, Fischer U
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
386 |
nm3 |
|
|
|
|
|
|
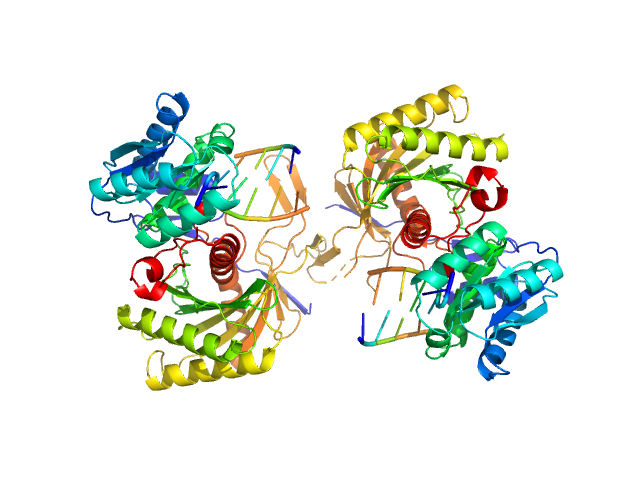
|
| Sample: |
Piwi protein AF_1318 dimer, 102 kDa Archaeoglobus fulgidus protein
5'-phosphorilated 14-mer DNA oligoduplex dimer, 9 kDa DNA
|
| Buffer: |
20 mM TrisHCl, pH 7.5, 5 mM MgCl2, 150 mM NaCl and 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 6
|
Prokaryotic Argonaute from Archaeoglobus fulgidus interacts with DNA as a homodimer.
Sci Rep 11(1):4518 (2021)
Golovinas E, Rutkauskas D, Manakova E, Jankunec M, Silanskas A, Sasnauskas G, Zaremba M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
158 |
nm3 |
|
|
|
|
|
|
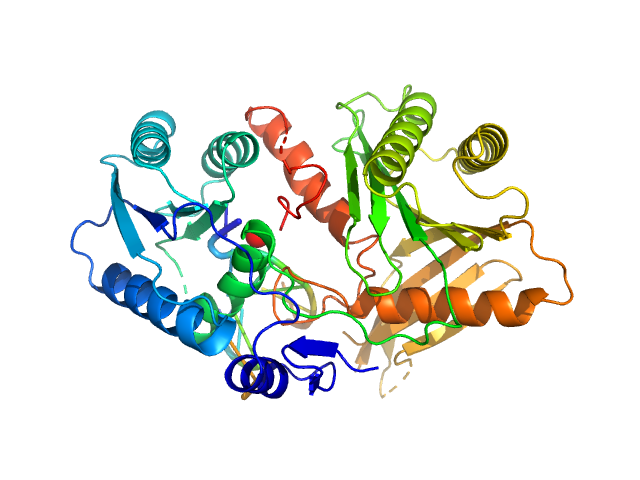
|
| Sample: |
Piwi protein AF_1318 delta (296-303) mutant monomer, 50 kDa Archaeoglobus fulgidus protein
5'-phosphorilated 14-mer DNA oligoduplex dimer, 9 kDa DNA
|
| Buffer: |
20 mM TrisHCl, pH 7.5, 5 mM MgCl2, 150 mM NaCl and 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 6
|
Prokaryotic Argonaute from Archaeoglobus fulgidus interacts with DNA as a homodimer.
Sci Rep 11(1):4518 (2021)
Golovinas E, Rutkauskas D, Manakova E, Jankunec M, Silanskas A, Sasnauskas G, Zaremba M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
109 |
nm3 |
|
|
|
|
|
|
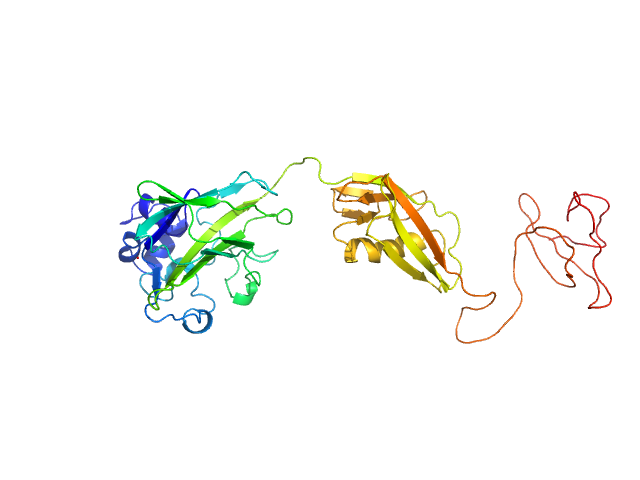
|
| Sample: |
Chitin-binding protein CbpD monomer, 39 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
15 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Oct 1
|
The lytic polysaccharide monooxygenase CbpD promotes Pseudomonas aeruginosa virulence in systemic infection.
Nat Commun 12(1):1230 (2021)
Askarian F, Uchiyama S, Masson H, Sørensen HV, Golten O, Bunæs AC, Mekasha S, Røhr ÅK, Kommedal E, Ludviksen JA, Arntzen MØ, Schmidt B, Zurich RH, van Sorge NM, Eijsink VGH, Krengel U, Mollnes TE, Lewis NE, Nizet V, Vaaje-Kolstad G
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
64131 |
nm3 |
|
|
|
|
|
|
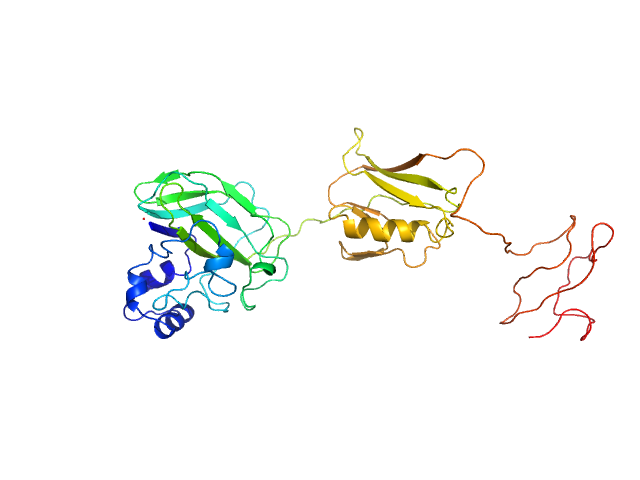
|
| Sample: |
Chitin-binding protein CbpD monomer, 39 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
15 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar with InCoatec Cu microsource, RECX, University of Oslo on 2019 Jul 19
|
The lytic polysaccharide monooxygenase CbpD promotes Pseudomonas aeruginosa virulence in systemic infection.
Nat Commun 12(1):1230 (2021)
Askarian F, Uchiyama S, Masson H, Sørensen HV, Golten O, Bunæs AC, Mekasha S, Røhr ÅK, Kommedal E, Ludviksen JA, Arntzen MØ, Schmidt B, Zurich RH, van Sorge NM, Eijsink VGH, Krengel U, Mollnes TE, Lewis NE, Nizet V, Vaaje-Kolstad G
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Chitin-binding protein CbpD monomer, 39 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
15 mM Tris-HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar with InCoatec Cu microsource, RECX, University of Oslo on 2019 Aug 14
|
The lytic polysaccharide monooxygenase CbpD promotes Pseudomonas aeruginosa virulence in systemic infection.
Nat Commun 12(1):1230 (2021)
Askarian F, Uchiyama S, Masson H, Sørensen HV, Golten O, Bunæs AC, Mekasha S, Røhr ÅK, Kommedal E, Ludviksen JA, Arntzen MØ, Schmidt B, Zurich RH, van Sorge NM, Eijsink VGH, Krengel U, Mollnes TE, Lewis NE, Nizet V, Vaaje-Kolstad G
|
| RgGuinier |
3.5 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
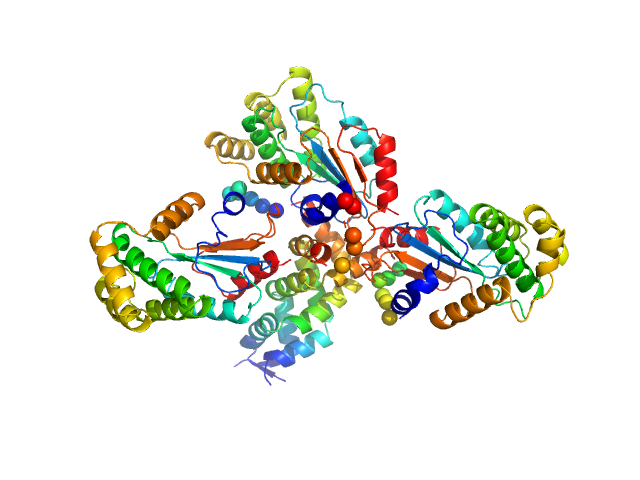
| Sample: |
Hypothetical exported protein trimer, 83 kDa Salmonella typhimurium (strain … protein
|
| Buffer: |
25 mM TRIS, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 14
|
Salmonella enterica BcfH is a trimeric thioredoxin-like bifunctional enzyme with both thiol oxidase and disulfide isomerase activities.
Antioxid Redox Signal (2021)
Subedi P, Paxman JJ, Wang G, Hor L, Hong Y, Verderosa AD, Whitten AE, Panjikar S, Santos-Martin CF, Martin JL, Totsika M, Heras B
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
|
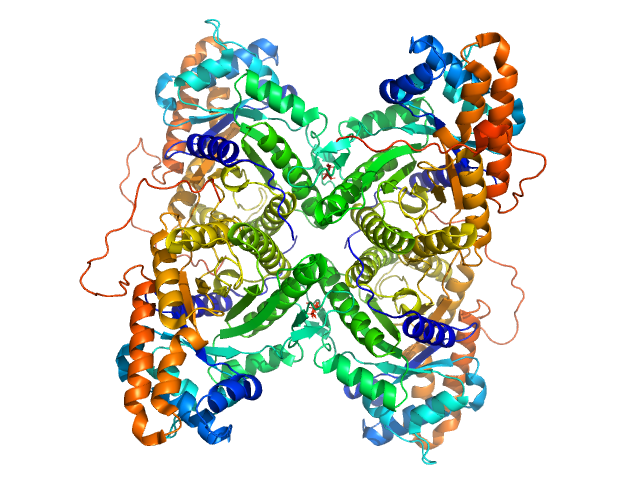
| Sample: |
Fructose-bisphosphate aldolase A tetramer, 157 kDa Oryctolagus cuniculus protein
|
| Buffer: |
20 mM HEPES, pH: 7 |
| Experiment: |
SAXS
data collected at Xenocs BioXolver L with MetalJet, Département de Biochimie, Université de Montréal on 2019 Jun 14
|
Rabbit muscle fructose-1,6-bisphosphate aldolase K229M mutant
Normand Cyr
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phage-encoded SAM lyase Svi3-3 (including N-terminal His6-tag and Tev cleavage site) trimer, 56 kDa Unknown environmental phage protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Dec 9
|
Structure and mechanism of a phage-encoded SAM lyase revises catalytic function of enzyme family.
Elife 10 (2021)
Guo X, Söderholm A, Kanchugal P S, Isaksen GV, Warsi O, Eckhard U, Trigüis S, Gogoll A, Jerlström-Hultqvist J, Åqvist J, Andersson DI, Selmer M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
85 |
nm3 |
|
|