|
|
|
|
|
| Sample: |
Endo-beta-N-acetylglucosaminidase H dimer, 61 kDa Streptomyces plicatus protein
|
| Buffer: |
20 mM Tris-HCl, 50 mM NaCl, 5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Nov 4
|
SAXS studies of X-ray induced disulfide bond damage: Engineering high-resolution insight from a low-resolution technique
PLOS ONE 15(11):e0239702 (2020)
Stachowski T, Snell M, Snell E, Boggon T
|
| RgGuinier |
2.7 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphoprotein monomer, 45 kDa Nipah henipavirus protein
|
| Buffer: |
20 mM Tris-HCl, 0.3 M NaCl, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 3
|
Ensemble description of the intrinsically disordered N-terminal domain of the Nipah virus P/V protein from combined NMR and SAXS.
Sci Rep 10(1):19574 (2020)
Schiavina M, Salladini E, Murrali MG, Tria G, Felli IC, Pierattelli R, Longhi S
|
| RgGuinier |
6.2 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|
|
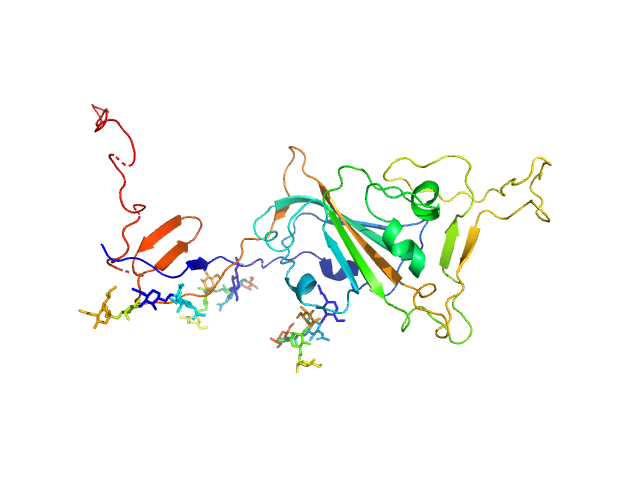
| Sample: |
Spike glycoprotein (ACE2 receptor binding domain) monomer, 29 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM Tris 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 May 1
|
Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2
Nature Communications 11(1) (2020)
Custódio T, Das H, Sheward D, Hanke L, Pazicky S, Pieprzyk J, Sorgenfrei M, Schroer M, Gruzinov A, Jeffries C, Graewert M, Svergun D, Dobrev N, Remans K, Seeger M, McInerney G, Murrell B, Hällberg B, Löw C
|
| RgGuinier |
3.0 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Apoferritin light chain 24-mer, 479 kDa Equus caballus protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 2% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Apr 5
|
Adding Size Exclusion Chromatography (SEC) and Light Scattering (LS) Devices to Obtain High-Quality Small Angle X-Ray Scattering (SAXS) Data
Crystals 10(11):975 (2020)
Graewert M, Da Vela S, Gräwert T, Molodenskiy D, Blanchet C, Svergun D, Jeffries C
|
| RgGuinier |
5.4 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
679 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Menisporopsin singlet acyl carrier protein-thioesterase monomer, 45 kDa Menisporopsin theobromae BCC … protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 25
|
SAXS reveals highly flexible interdomain linkers of tandem acyl carrier protein–thioesterase domains from a fungal nonreducing polyketide synthase
FEBS Letters (2020)
Bunnak W, Winter A, Lazarus C, Crump M, Race P, Wattana‐Amorn P
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Menisporopsin doublet Acyl Carrier Protein-Thioesterase monomer, 58 kDa Menisporopsis theobromae BCC … protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jan 25
|
SAXS reveals highly flexible interdomain linkers of tandem acyl carrier protein–thioesterase domains from a fungal nonreducing polyketide synthase
FEBS Letters (2020)
Bunnak W, Winter A, Lazarus C, Crump M, Race P, Wattana‐Amorn P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.3 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
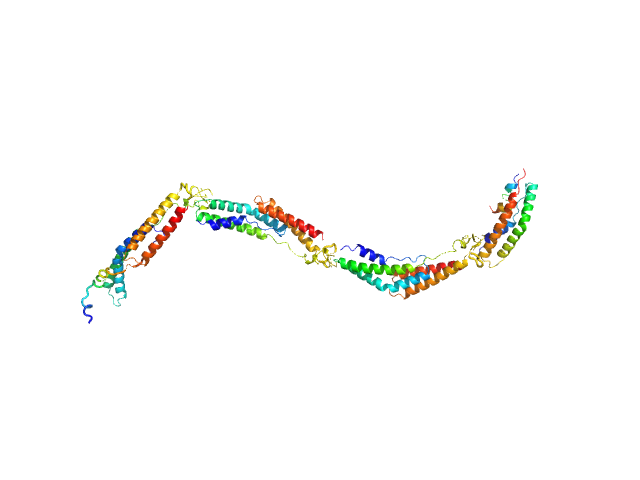
| Sample: |
Extracellular matrix binding protein F-repeats, 70 kDa Staphylococcus epidermidis protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 15
|
A Giant Extracellular Matrix Binding Protein of Staphylococcus epidermidis Binds Surface-Immobilized Fibronectin via a Novel Mechanism.
mBio 11(5) (2020)
Büttner H, Perbandt M, Kohler T, Kikhney A, Wolters M, Christner M, Heise M, Wilde J, Weißelberg S, Both A, Betzel C, Hammerschmidt S, Svergun D, Aepfelbacher M, Rohde H
|
| RgGuinier |
7.0 |
nm |
| Dmax |
28.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Extracellular matrix binding protein FG-repeats, 85 kDa Staphylococcus epidermidis protein
|
| Buffer: |
PBS, 136.5 mM NaCl, 2.65 mM KCl, 8.3 mM Na2HPO4, 2.65 mM KH2PO4, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jun 14
|
A Giant Extracellular Matrix Binding Protein of Staphylococcus epidermidis Binds Surface-Immobilized Fibronectin via a Novel Mechanism.
mBio 11(5) (2020)
Büttner H, Perbandt M, Kohler T, Kikhney A, Wolters M, Christner M, Heise M, Wilde J, Weißelberg S, Both A, Betzel C, Hammerschmidt S, Svergun D, Aepfelbacher M, Rohde H
|
| RgGuinier |
10.7 |
nm |
| Dmax |
40.0 |
nm |
|
|
|
|
|
|
|
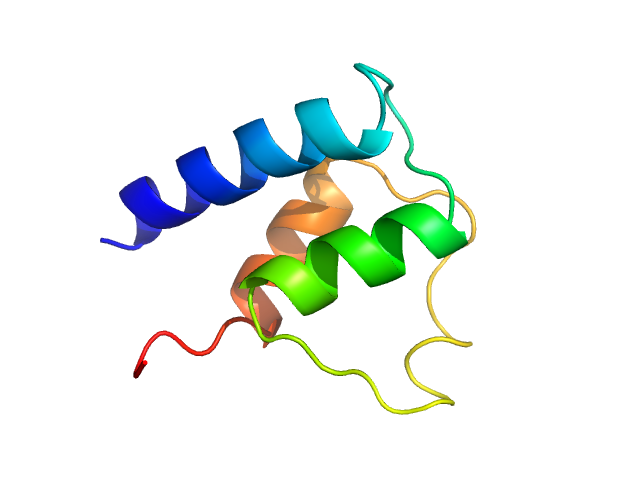
| Sample: |
Myosin essential light chain monomer, 9 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Oct 25
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Myosin essential light chain monomer, 16 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jun 30
|
Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol 3(1):568 (2020)
Pazicky S, Dhamotharan K, Kaszuba K, Mertens HDT, Gilberger T, Svergun D, Kosinski J, Weininger U, Löw C
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
23 |
nm3 |
|
|