|
|
|
|
|
| Sample: |
Bcl-2-like protein FPV039 monomer, 17 kDa Fowlpox virus protein
Uncharacterized protein (BAK1) monomer, 3 kDa Gallus gallus protein
|
| Buffer: |
20 mM trisodium citrate pH, 200 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Oct 2
|
Structural basis of apoptosis inhibition by the fowlpox virus protein FPV039.
J Biol Chem 292(22):9010-9021 (2017)
Anasir MI, Caria S, Skinner MA, Kvansakul M
|
|
|
|
|
|
|
|
| Sample: |
Poly-deoxythymidine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 Apr 1
|
Visualizing single-stranded nucleic acids in solution.
Nucleic Acids Res 45(9):e66 (2017)
Plumridge A, Meisburger SP, Pollack L
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.7 |
nm |
|
|
|
|
|
|
|
| Sample: |
Poly-deoxyadenosine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Apr 1
|
Visualizing single-stranded nucleic acids in solution.
Nucleic Acids Res 45(9):e66 (2017)
Plumridge A, Meisburger SP, Pollack L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
|
|
|
|
|
|
|
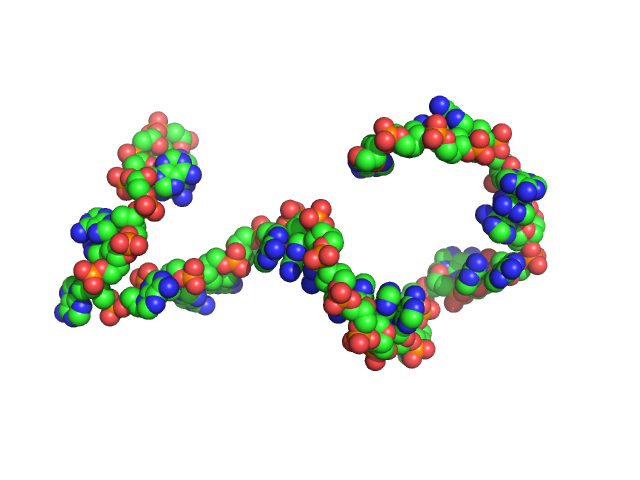
| Sample: |
Poly-deoxyadenosine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1mM Na MOPS, 100mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Apr 1
|
The impact of base stacking on the conformations and electrostatics of single-stranded DNA.
Nucleic Acids Res 45(7):3932-3943 (2017)
Plumridge A, Meisburger SP, Andresen K, Pollack L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
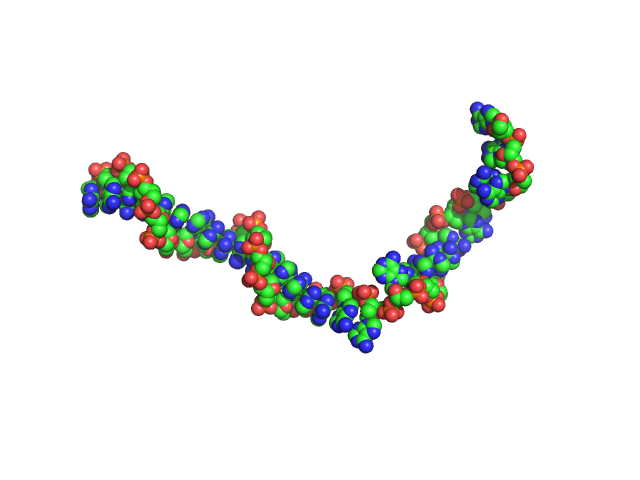
| Sample: |
Poly-deoxyadenosine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1mM Na MOPS, 200mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Apr 1
|
The impact of base stacking on the conformations and electrostatics of single-stranded DNA.
Nucleic Acids Res 45(7):3932-3943 (2017)
Plumridge A, Meisburger SP, Andresen K, Pollack L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
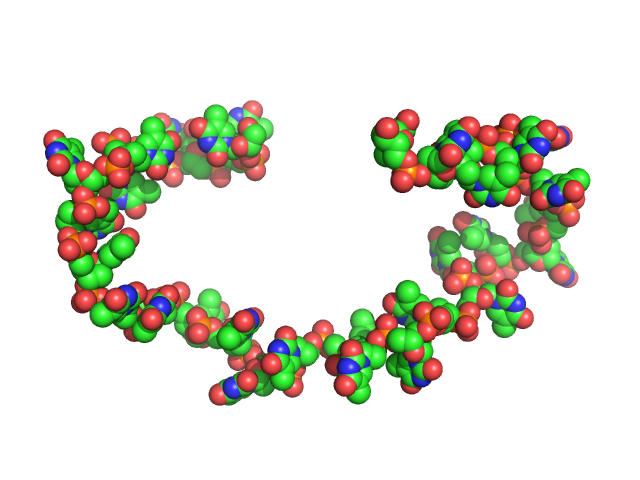
|
| Sample: |
Poly-deoxythymidine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1mM Na MOPS, 100mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Apr 1
|
The impact of base stacking on the conformations and electrostatics of single-stranded DNA.
Nucleic Acids Res 45(7):3932-3943 (2017)
Plumridge A, Meisburger SP, Andresen K, Pollack L
|
|
|
|
|
|
|
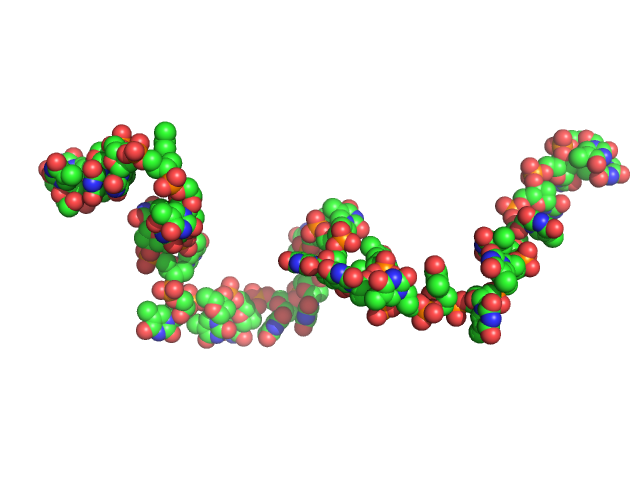
|
| Sample: |
Poly-deoxythymidine (30mer) monomer, 9 kDa DNA
|
| Buffer: |
1mM Na MOPS, 200mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Apr 1
|
The impact of base stacking on the conformations and electrostatics of single-stranded DNA.
Nucleic Acids Res 45(7):3932-3943 (2017)
Plumridge A, Meisburger SP, Andresen K, Pollack L
|
|
|
|
|
|
|
|
| Sample: |
ESX-5 type VII secretion system protein EccC5 monomer, 142 kDa Mycobacterium xenopi RIVM700367 protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 5%(v/v) Glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Jul 15
|
Structure of the mycobacterial ESX-5 type VII secretion system membrane complex by single-particle analysis.
Nat Microbiol 2:17047 (2017)
Beckham KS, Ciccarelli L, Bunduc CM, Mertens HD, Ummels R, Lugmayr W, Mayr J, Rettel M, Savitski MM, Svergun DI, Bitter W, Wilmanns M, Marlovits TC, Parret AH, Houben EN
|
| RgGuinier |
6.2 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
255 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Type-2 restriction enzyme AgeI monomer, 31 kDa Thalassobius gelatinovorus protein
|
| Buffer: |
10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM CaCl₂, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 14
|
Restriction endonuclease AgeI is a monomer which dimerizes to cleave DNA.
Nucleic Acids Res 45(6):3547-3558 (2017)
Tamulaitiene G, Jovaisaite V, Tamulaitis G, Songailiene I, Manakova E, Zaremba M, Grazulis S, Xu SY, Siksnys V
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
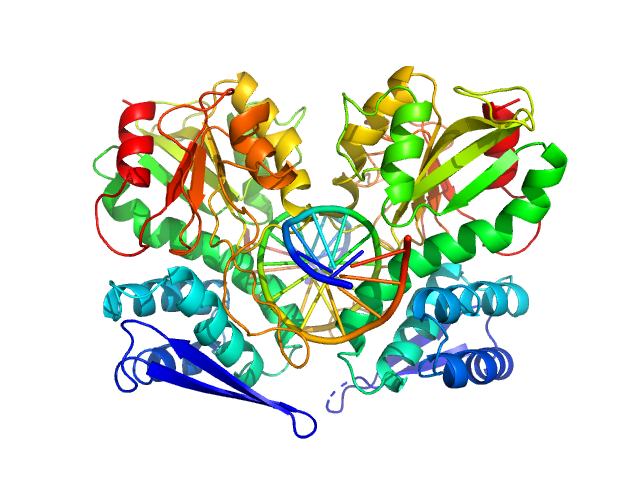
|
| Sample: |
Type-2 restriction enzyme AgeI dimer, 61 kDa Thalassobius gelatinovorus protein
Cognate DNA oligoduplex with 5'-T overhang dimer, 8 kDa DNA
|
| Buffer: |
10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM CaCl₂, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 14
|
Restriction endonuclease AgeI is a monomer which dimerizes to cleave DNA.
Nucleic Acids Res 45(6):3547-3558 (2017)
Tamulaitiene G, Jovaisaite V, Tamulaitis G, Songailiene I, Manakova E, Zaremba M, Grazulis S, Xu SY, Siksnys V
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
69 |
nm3 |
|
|