|
|
|
|

|
| Sample: |
TnpA transposase dimer, 234 kDa Bacillus thuringiensis serovar … protein
|
| Buffer: |
50 mM HEPES, 200 mM NaCl, 100 mM L-Arg HCL, pH: 7.9 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Nov 2
|
AFM-based force spectroscopy unravels stepwise formation of the DNA transposition complex in the widespread Tn3 family mobile genetic elements.
Nucleic Acids Res (2023)
Fernandez M, Shkumatov AV, Liu Y, Stulemeijer C, Derclaye S, Efremov RG, Hallet B, Alsteens D
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
480 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform P3 of Phosphoprotein dimer, 55 kDa Rabies virus (strain … protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 26
|
Structural insights into the multifunctionality of rabies virus P3 protein.
Proc Natl Acad Sci U S A 120(14):e2217066120 (2023)
Sethi A, Rawlinson SM, Dubey A, Ang CS, Choi YH, Yan F, Okada K, Rozario AM, Brice AM, Ito N, Williamson NA, Hatters DM, Bell TDM, Arthanari H, Moseley GW, Gooley PR
|
| RgGuinier |
3.7 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Attenuated derivative P3 of Phosphoprotein dimer, 55 kDa protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 26
|
Structural insights into the multifunctionality of rabies virus P3 protein.
Proc Natl Acad Sci U S A 120(14):e2217066120 (2023)
Sethi A, Rawlinson SM, Dubey A, Ang CS, Choi YH, Yan F, Okada K, Rozario AM, Brice AM, Ito N, Williamson NA, Hatters DM, Bell TDM, Arthanari H, Moseley GW, Gooley PR
|
| RgGuinier |
4.0 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform P3 of Phosphoprotein Nish P3 N226H dimer, 55 kDa protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 26
|
Structural insights into the multifunctionality of rabies virus P3 protein.
Proc Natl Acad Sci U S A 120(14):e2217066120 (2023)
Sethi A, Rawlinson SM, Dubey A, Ang CS, Choi YH, Yan F, Okada K, Rozario AM, Brice AM, Ito N, Williamson NA, Hatters DM, Bell TDM, Arthanari H, Moseley GW, Gooley PR
|
| RgGuinier |
4.3 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
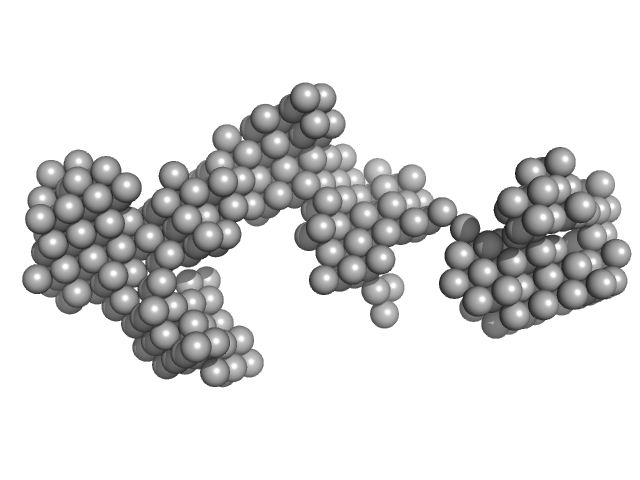
|
| Sample: |
Japanese encephaltitis 5' TR monomer, 74 kDa Japanese encephalitis virus RNA
|
| Buffer: |
10 mM Bis-tris, 100 mM NaCl, 15 mM KCl 15 mM MgCl2, 10% glycerol, pH: 5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 22
|
Investigating RNA-RNA interactions through computational and biophysical analysis.
Nucleic Acids Res (2023)
Mrozowich T, Park SM, Waldl M, Henrickson A, Tersteeg S, Nelson CR, De Klerk A, Demeler B, Hofacker IL, Wolfinger MT, Patel TR
|
| RgGuinier |
7.0 |
nm |
| Dmax |
22.4 |
nm |
| VolumePorod |
356 |
nm3 |
|
|
|
|
|
|
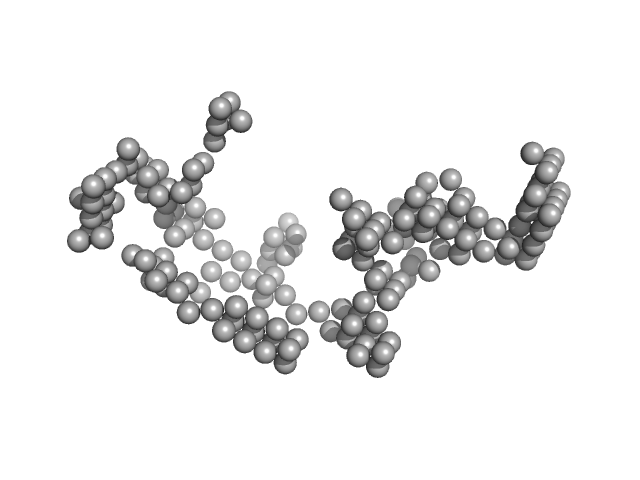
|
| Sample: |
Japanese encephalitis virus 3' UTR monomer, 186 kDa Japanese encephalitis virus RNA
|
| Buffer: |
10 mM Bis-tris, 100 mM NaCl, 15 mM KCl 15 mM MgCl2, 10% glycerol, pH: 5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 22
|
Investigating RNA-RNA interactions through computational and biophysical analysis.
Nucleic Acids Res (2023)
Mrozowich T, Park SM, Waldl M, Henrickson A, Tersteeg S, Nelson CR, De Klerk A, Demeler B, Hofacker IL, Wolfinger MT, Patel TR
|
| RgGuinier |
11.3 |
nm |
| Dmax |
34.6 |
nm |
| VolumePorod |
2900 |
nm3 |
|
|
|
|
|
|
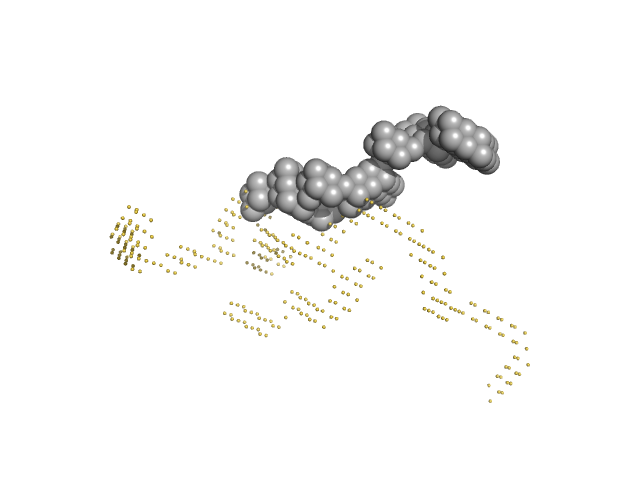
|
| Sample: |
Japanese encephalitis virus 5' TR and 3' UTR complex monomer, 260 kDa Japanese encephalitis virus RNA
|
| Buffer: |
10 mM Bis-tris, 100 mM NaCl, 15 mM KCl 15 mM MgCl2, 10% glycerol, pH: 5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 22
|
Investigating RNA-RNA interactions through computational and biophysical analysis.
Nucleic Acids Res (2023)
Mrozowich T, Park SM, Waldl M, Henrickson A, Tersteeg S, Nelson CR, De Klerk A, Demeler B, Hofacker IL, Wolfinger MT, Patel TR
|
| RgGuinier |
12.8 |
nm |
| Dmax |
40.0 |
nm |
| VolumePorod |
4550 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human dystrophin central domain R4-15 fragment monomer, 150 kDa protein
|
| Buffer: |
NaP 20 mM, NaCl 300 mM, EDTA 1 mM, Glycérol 2%, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 May 26
|
Dystrophin SAXS data
Raphael Dos Santos Morais
|
| RgGuinier |
9.7 |
nm |
| Dmax |
47.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
Human dystrophin central domain R16-24 fragment monomer, 127 kDa protein
|
| Buffer: |
NaP 20 mM, NaCl 300 mM, EDTA 1 mM, Glycérol 2%, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Dec 17
|
Dystrophin SAXS data
Raphael Dos Santos Morais
|
| RgGuinier |
9.3 |
nm |
| Dmax |
34.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Human dystrophin central domain R16-24 del45-55 fragment monomer, 58 kDa protein
|
| Buffer: |
NaP 20 mM, NaCl 300 mM, EDTA 1 mM, Glycérol 2%, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Sep 28
|
Dystrophin SAXS data
Raphael Dos Santos Morais
|
| RgGuinier |
4.7 |
nm |
| Dmax |
18.3 |
nm |
|
|