|
|
|
|
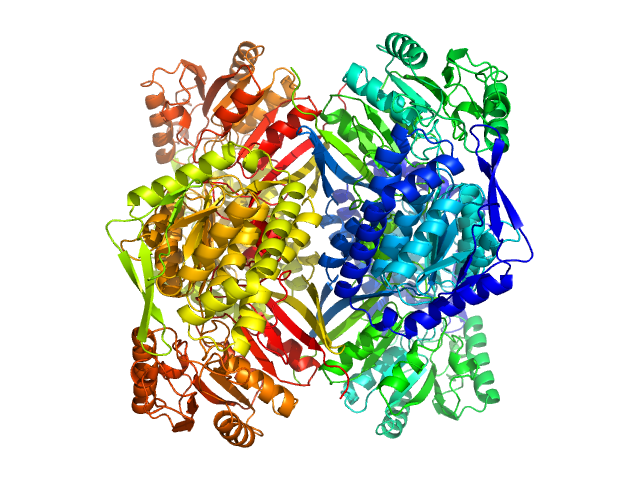
|
| Sample: |
2-aminomuconic 6-semialdehyde dehydrogenase tetramer, 215 kDa Pseudomonas sp. protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, pH: 7
|
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Feb 1
|
The tetrameric assembly of 2-aminomuconic 6-semialdehyde dehydrogenase is a functional requirement of cofactor NAD(+) binding.
Environ Microbiol 24(7):2994-3012 (2022)
Shi Q, Chen Y, Li X, Dong H, Chen C, Zhong Z, Yang C, Liu G, Su D
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
326 |
nm3 |
|
|
|
|
|
|
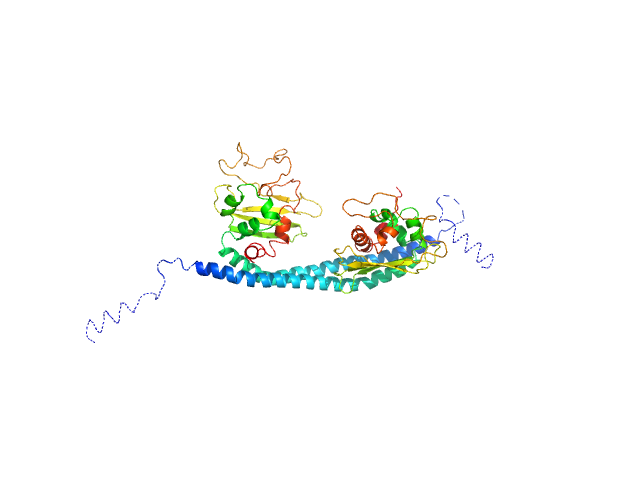
|
| Sample: |
SET nuclear proto-oncogene dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
Sodium phosphate buffer, pH: 6.3
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 11
|
PP2A is activated by cytochrome c upon formation of a diffuse encounter complex with SET/TAF-Iβ
Computational and Structural Biotechnology Journal (2022)
Casado-Combreras M, Rivero-Rodríguez F, Elena-Real C, Molodenskiy D, Díaz-Quintana A, Martinho M, Gerbaud G, González-Arzola K, Velázquez-Campoy A, Svergun D, Belle V, De la Rosa M, Díaz-Moreno I
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
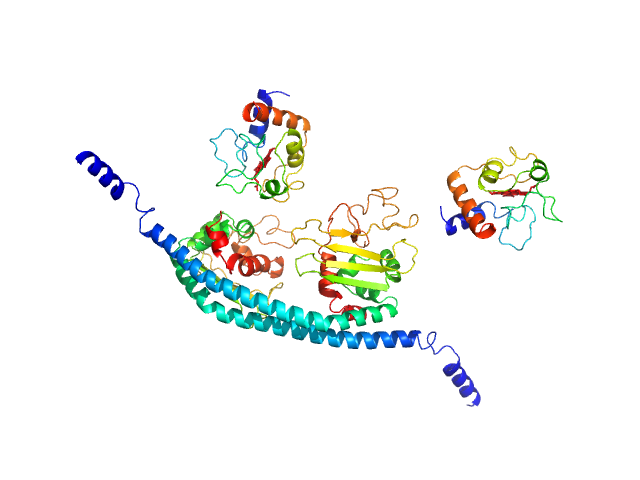
|
| Sample: |
Cytochrome c dimer, 24 kDa Homo sapiens protein
SET nuclear proto-oncogene dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
Sodium phosphate buffer, pH: 6.3
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 11
|
PP2A is activated by cytochrome c upon formation of a diffuse encounter complex with SET/TAF-Iβ
Computational and Structural Biotechnology Journal (2022)
Casado-Combreras M, Rivero-Rodríguez F, Elena-Real C, Molodenskiy D, Díaz-Quintana A, Martinho M, Gerbaud G, González-Arzola K, Velázquez-Campoy A, Svergun D, Belle V, De la Rosa M, Díaz-Moreno I
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
|
|
|
|
|
|
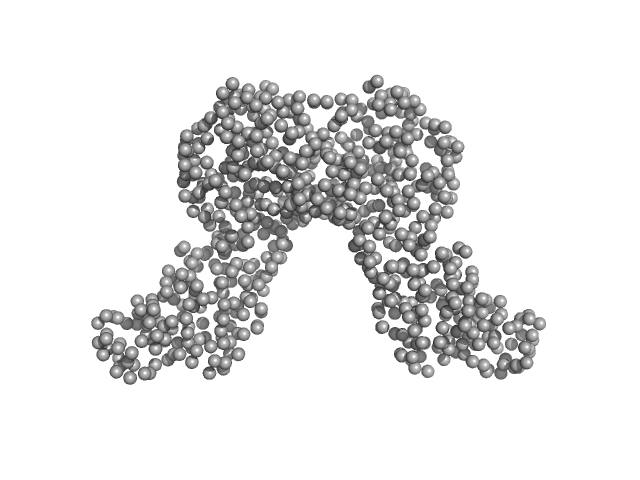
|
| Sample: |
Riboflavin biosynthesis protein RibD dimer, 80 kDa Acinetobacter baumannii protein
|
| Buffer: |
150 mM NaCl, 10 mM Tris, 1 mM DTT, 5% v/v glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 3
|
Recombinant AcnB, NrdR and RibD of Acinetobacter baumannii and their potential interaction with DNA adenine methyltransferase AamA.
Protein Expr Purif :106134 (2022)
Weber K, Doellinger J, Jeffries CM, Wilharm G
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aconitate hydratase B monomer, 96 kDa Acinetobacter baumannii protein
|
| Buffer: |
100 mM NaCl, 20 mM Tris, 1 mM DTT, 3% v/v glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 3
|
Recombinant AcnB, NrdR and RibD of Acinetobacter baumannii and their potential interaction with DNA adenine methyltransferase AamA.
Protein Expr Purif :106134 (2022)
Weber K, Doellinger J, Jeffries CM, Wilharm G
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
|
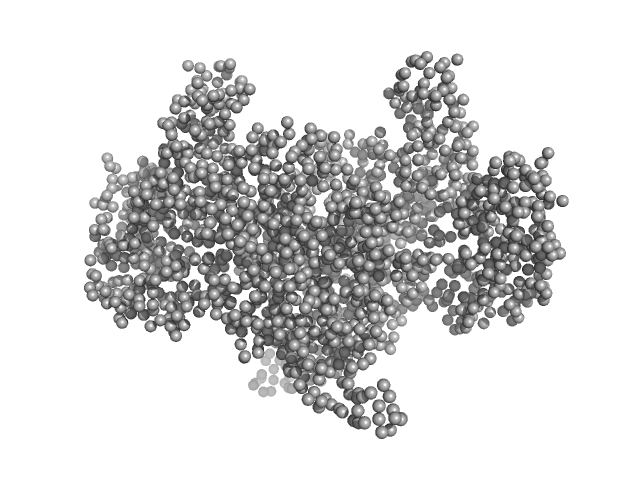
| Sample: |
Aconitate hydratase B , 96 kDa Acinetobacter baumannii protein
|
| Buffer: |
100 mM NaCl, 20 mM Tris, 1 mM DTT, 3% v/v glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 3
|
Recombinant AcnB, NrdR and RibD of Acinetobacter baumannii and their potential interaction with DNA adenine methyltransferase AamA.
Protein Expr Purif :106134 (2022)
Weber K, Doellinger J, Jeffries CM, Wilharm G
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
|
|
|
|
|
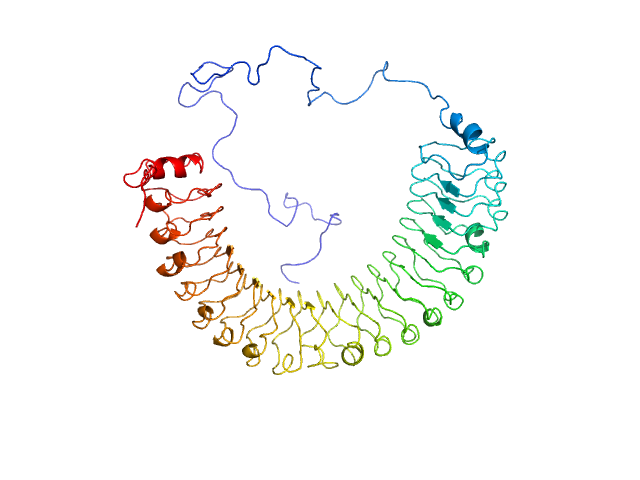
| Sample: |
Transcriptional repressor NrdR , 19 kDa Acinetobacter baumannii (strain … protein
|
| Buffer: |
100 mM NaCl, 20 mM Tris, 1 mM DTT, 3% v/v glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Dec 3
|
Recombinant AcnB, NrdR and RibD of Acinetobacter baumannii and their potential interaction with DNA adenine methyltransferase AamA.
Protein Expr Purif :106134 (2022)
Weber K, Doellinger J, Jeffries CM, Wilharm G
|
| RgGuinier |
19.9 |
nm |
| Dmax |
92.6 |
nm |
|
|
|
|
|
|
|
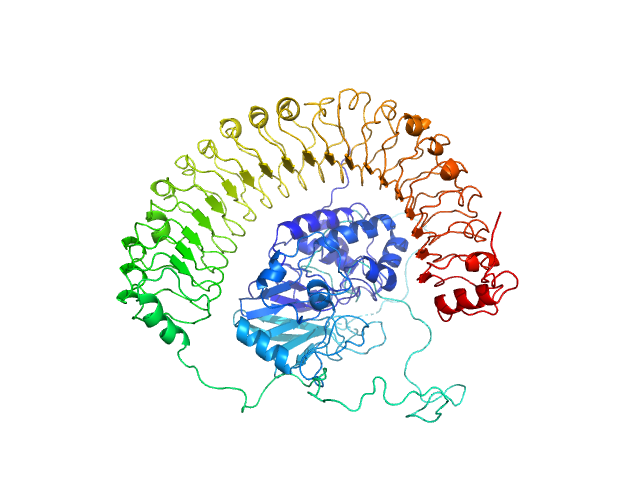
| Sample: |
Leucine-rich repeat protein SHOC-2 monomer, 65 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris pH 7.5, 100 mM NaCl, 1 mM TCEP, 1 mM MnCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jun 20
|
Structural basis for SHOC2 modulation of RAS signalling.
Nature (2022)
Liau NPD, Johnson MC, Izadi S, Gerosa L, Hammel M, Bruning JM, Wendorff TJ, Phung W, Hymowitz SG, Sudhamsu J
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|
|
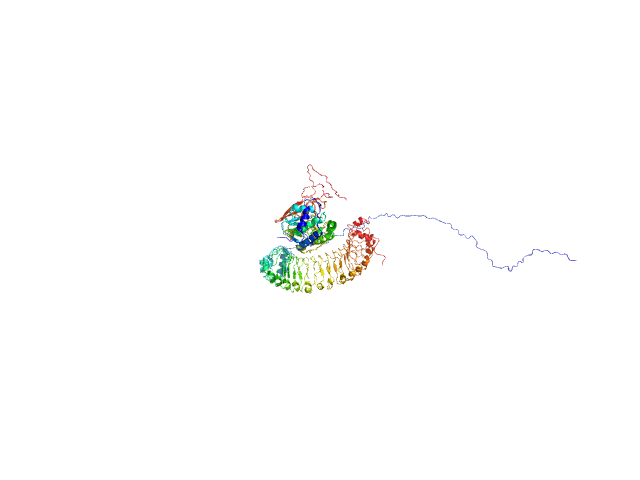
| Sample: |
Leucine-rich repeat protein SHOC-2 monomer, 65 kDa Homo sapiens protein
Serine/threonine-protein phosphatase PP1-gamma catalytic subunit monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris pH 7.5, 100 mM NaCl, 1 mM TCEP, 1 mM MnCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 20
|
Structural basis for SHOC2 modulation of RAS signalling.
Nature (2022)
Liau NPD, Johnson MC, Izadi S, Gerosa L, Hammel M, Bruning JM, Wendorff TJ, Phung W, Hymowitz SG, Sudhamsu J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Leucine-rich repeat protein SHOC-2 monomer, 65 kDa Homo sapiens protein
Serine/threonine-protein phosphatase PP1-gamma catalytic subunit monomer, 37 kDa Homo sapiens protein
GTPase KRas monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris pH 7.5, 100 mM NaCl, 1 mM TCEP, 1 mM MnCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Jul 20
|
Structural basis for SHOC2 modulation of RAS signalling.
Nature (2022)
Liau NPD, Johnson MC, Izadi S, Gerosa L, Hammel M, Bruning JM, Wendorff TJ, Phung W, Hymowitz SG, Sudhamsu J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
156 |
nm3 |
|
|